THE YEAST TWO-HYBRID ASSAY: AN EXERCISE IN EXPERIMENTAL ELOQUENCE
(August, 2003)
Once upon a time, it was believed that proteins were isolated entities, floating in the cytosol and, for the most part, acting independently of surrounding proteins. Proteins were thought to diffuse freely, and reactions occurred as a result of proteins A and B randomly colliding with one another. Today we know this picture to be far too simplistic to account for the complex processes that all coalesce to become ‘life’. Instead, the majority of cellular phenomena are carried out by protein ‘machines’, or aggregates of ten or more proteins [1]. These protein-protein interactions are critical to all cellular processes, and understanding them is key to understanding any biological system. One technique that can be used to study protein-protein interactions is the “yeast two hybrid” system.
Yeasty goodness
A protein is composed of modules or domains, which are individually folded units within the same polypeptide (protein) chain. The presence of these individual domains allow the same protein to perform different functions. The yeast two-hybrid technique uses two protein domains that have specific functions: a DNA-binding domain (BD), that is capable of binding to DNA, and an activation domain (AD), that is capable of activating transcription of the DNA.
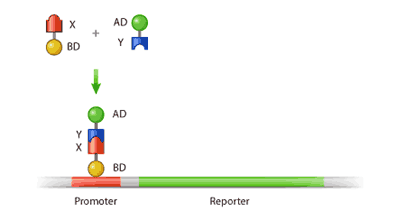
Both of these domains are required for transcription, whereby DNA is copied in the form of mRNA, which is later translated into protein. In order for DNA to be transcribed, it requires a protein called a transcriptional activator (TA). This protein binds to the “promoter”, a region situated upstream from the gene (coding region of the DNA) that serves as a docking site for the transcriptional protein (Figure 1). Once the TA has bound to the promoter, it is then able to activate transcription via its activation domain. Hence, the activity of a TA requires both a DNA binding domain and an activation domain. If either of these domains is absent, then transcription of the gene will fail.

Furthermore, the binding domain and the activation domain do not necessarily have to be on the same protein. In fact, a protein with a DNA binding domain can activate transcription when simply bound to another protein containing an activation domain; this principle forms the basis for the yeast two-hybrid technique2.
In the two-hybrid assay, two fusion proteins are created: the protein of interest (X), which is constructed to have a DNA binding domain attached to its N-terminus, and its potential binding partner (Y), which is fused to an activation domain. If protein X interacts with protein Y, the binding of these two will form an intact and functional transcriptional activator [2]. This newly formed transcriptional activator will then go on to transcribe a reporter gene, which is simply a gene whose protein product can be easily detected and measured. In this way, the amount of the reporter produced can be used as a measure of interaction between our protein of interest and its potential partner (Figure 2).
The Recipe for Successful Interactions
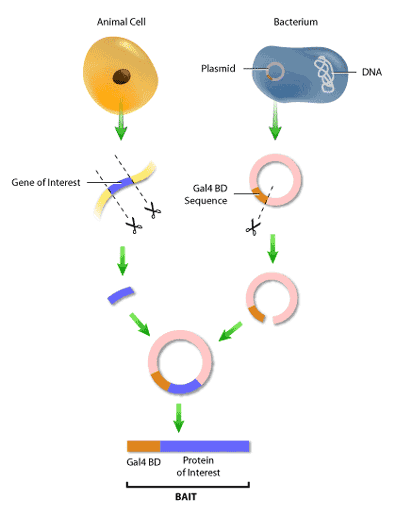
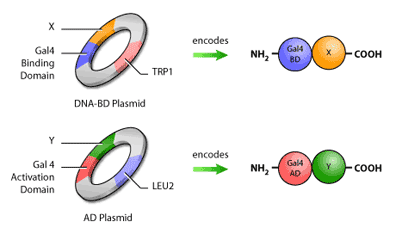
First, it is necessary to construct the ‘bait’ and ‘hunter’ fusion proteins. The ‘bait’ fusion protein is the protein of interest (or ‘bait’) linked to the GAL4 binding domain, or GAL4 BD. This is done by inserting the segment of DNA encoding the bait into a plasmid, which is a small circular molecule of double-stranded DNA that occurs naturally in both bacteria and yeast. This plasmid will also have inserted in it a segment of Gal4 BD DNA next to the site of bait DNA insertion. Therefore, when the DNA from the plasmid is transcribed and converted to protein, the bait will now have a binding domain attached to its end (Figure 3). The same procedure is used to construct the ‘hunter’ protein, where the potential binding partner is fused to the GAL4 AD.
In addition to having the fusion proteins encoded for, these plasmids will also contain selection genes, or genes encoding proteins that contribute to a cell’s survival in a particular environment. An example of a selection gene is one encoding antibiotic resistance; when antibiotics are introduced, only cells with the antibiotic resistance gene will survive. Yeast two-hybrid assays typically use selection genes encoding proteins capable of synthesizing amino acids such as histidine, leucine and tryptophan (Figure 4).
Once the plasmids have been constructed, they must next be introduced into a host yeast cell by a process called “transfection”. In this process, the outer-membrane of a yeast cell is disturbed by a physical method, such as sonification or chemical disruption. This disruption produces holes that are large enough for the plasmid to enter, and in this way, the plasmids can cross the membrane and enter the cell (Figure 5).
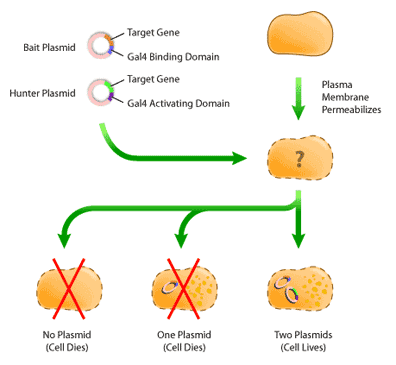
Once the cells have been transfected, it is necessary to isolate colonies that have both ‘bait’ and ‘hunter’ plasmids. This is because not every cell will have both plasmids cross their plasma membrane; some will have only one plasmid, while others will have none. Isolation of transfected cells involves identifying cells containing plasmids by virtue of their expressing the selection genes mentioned previously. After the cells have been transfected and allowed to recover for several days, they are then plated on minimal media, or media that is lacking one essential nutrient, such as tryptophan. The cells used for transfection are called auxotrophic mutants; these cells are deficient in producing nutrients required for their growth. By supplying the gene for the deficient nutrient in the ‘bait’ or ‘hunter’ plasmid, cells containing the plasmid are able to survive on the minimal media, whereas untransfected cells cannot (Figure 5). Selection in this way occurs in two rounds: first on one minimal media plate, to select for the ‘bait’ plasmid, and then on another minimal media plate, to select for the ‘hunter’ [4].
Once inside the cell, if binding occurs between the hunter and the bait, transcriptional activity will be restored and will produce normal Gal4 activity. The reporter gene most commonly used in the Gal4 system is LacZ, an E. coli gene whose transcription causes cells to turn blue [4]. In this yeast system, the LacZ gene is inserted in the yeast DNA immediately after the Gal4 promoter, so that if binding occurs, LacZ is produced. Therefore, detecting interactions between bait and hunter simply requires identifying blue versus non-blue.
What Can I Do With My Very Own Yeast Two-Hybrid?
Generally the yeast two-hybrid assay can identify novel protein-protein interactions. By using a number of different proteins as potential binding partners, it is possible to detect interactions that were previously uncharacterized [3]. Secondly, the yeast two-hybrid assay can be used to characterize interactions already known to occur. Characterization could include determining which protein domains are responsible for the interaction, by using truncated proteins, or under what conditions interactions take place, by altering the intracellular environment.
The last and most recent application of the yeast two-hybrid involves manipulating protein-protein interactions in an attempt to understand its biological relevance. For example, many disorders arise due to mutations causing the protein to be non-functional, or have altered function. Such is the case of some cancers; a mutation in a pro-growth pathway does not allow for the binding of negative regulatory proteins, resulting in the pro-growth pathway never turning ‘off’. The yeast two-hybrid is one means of determining how mutation affects a protein’s interaction with other proteins. When a mutation is identified that affects binding, the significance of this mutation can be studied further by creating an organism that has this mutation and characterizing its phenotype.
Conclusion…
The yeast two-hybrid assay is an elegant means of investigating protein-protein interactions. A fairly new addition to the family of microbiological studies, these interactions have become increasingly important to our understanding of biological systems in the past few years. While isolation of a protein in an attempt to understand its function still has its place in biological research, we now understand that biological reactions do not occur in isolation. A protein is constantly interacting with other proteins in what we now know to be a delicate balance — to ignore this wealth of information would be to deny ourselves the opportunity to fully appreciate the stuff that life is made of.
References
1. Alberts, B. (1998). The Cell as a Collection of Protein Machines: Preparing the Next Generation of Molecular Biologists. Cell 92: 291-4.
2. Fields S, Song O.K. (1989). A novel genetic system to detect protein-protein interactions. Nature 340:245-6.
3. Fields S, Sternglanz R. (1994). The two-hybrid system: an assay for protein-protein interactions. Trends in Genetics 10:286-92.
4. Two-hybrid analysis of genetic regulatory networks — online protocol
Additional Reading
Bartel P, Fields S. (eds). (1997). The yeast two-hybrid system. New York: Oxford University Press.
(Art by Jiang Long – note that high res versions of image files available here)