MICROBES AND YOU: NORMAL FLORA
(August 2003)
Microbes are everywhere. They populate the air, the water, the soil, and have even evolved intimate relationships with plants and animals. Without microbes, life on earth would cease. This is due mainly to the essential roles microbes play in the systems that support life on earth, such as nutrient cycling and photosynthesis. Further, the physiology, nutrition and protection of plants and animals (including humans) is dependent on various relationships with microbes. This report will focus on the relationships between microbes and humans. And as we will see these relationships are key factors that determine whether or not we live healthy lives.
Microbes and You
You are covered in microorganisms! In fact, there are approximately 10 times as many prokaryotic cells (mainly bacteria) associated with your body than there are eukaryotic cells, but this is a good thing.
Microbes that colonize the human body during birth or shortly thereafter, remaining throughout life, are referred to as normal flora [1-2]. Normal flora can be found in many sites of the human body including the skin (especially the moist areas, such as the groin and between the toes), respiratory tract (particularly the nose), urinary tract, and the digestive tract (primarily the mouth and the colon). On the other hand, areas of the body such as the brain, the circulatory system and the lungs are intended to remain sterile (microbe free).
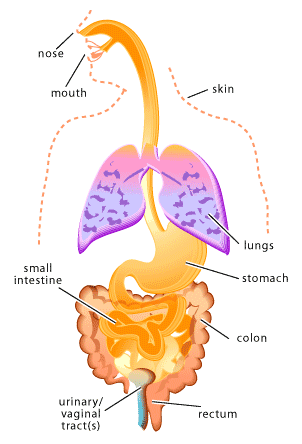
The human body provides many unique environments for different bacterial communities to live. In this context, scientists refer to the human body as the host. A positive host-microbe relationship is usually described as either mutualistic or commensalistic. In mutualism both the host and the microbe benefit. Which is in contract to commensalisms, where one partner of the relationship benefits (usually the microbe) and the other partner (usually the host) is neither benefited nor harmed. In many cases it may be difficult to establish whether a particular host-microbe relationship should be considered mutualistic or commensalistic, since scientists are only beginning to understand the role of normal flora in human health. In other words, individual microbes may be carrying out important functions within our bodies that we have not yet discovered. Just as host-microbe relationships can be positive or neutral, they can also be negative. Such a host-microbe relationship is usually described as parasitic or pathogenic. In a parasitic relationship the microbe benefits at the expense of the host and similarly in a pathogenic relationship the microbe causes damage to the host. In both cases the cost to the host can vary from slight to fatal.
Whether a host-microbe relationship is “positive” or “negative” depends on many factors. And in most cases the relationship will actually remain positive. The host provides a niche and nutrition for the colonizing microbe and the microbe occupies a space that a potential parasite or pathogen might otherwise colonize. In these cases microbial communities may even aid in digestion or synthesize nutrients for the host. However, life is not always perfect, and in certain situations good-standing members of your normal flora can cause disease or invading pathogens can displace them. The result will be disease. To illustrate some of these scenarios let’s take a closer look at microbial communities found in different areas of the human body.
Life on the Surface, the Skin
Human skin is not a particularly rich place for microbes to live. The skin surface is relatively dry, slightly acidic and the primary source of nutrition is dead cells. This is an environment that prevents the growth of many microorganisms, but a few have adapted to life on our skin.
Propionibacterium acnes is a Gram positive bacterium that inhabits the skin. P. acnes are anaerobes, so they lives in pores and glands where oxygen levels are lower. As the name implies P. acnes causes the common skin condition called acne. Although acne outbreaks can result in emotional and physical discomfort the infection is not life threatening. A point complemented by P. acnes performing an important role through occupying niches that might otherwise be colonized by more dangerous pathogens.
Another prominent member of the skin flora is Staphylococcus epidermidis. This is a highly adapted Gram positive bacterium that can survive at many sites throughout the body. S. epidermidis can cause life threatening disease in hospital patients when invasive medical devices such as catheters are used. In such cases, S. epidermidis form antibiotic resistant biofilms along the catheter and enter the bloodstream causing systemic infection that can be fatal. Under this scenario S. epidermidis would be considered an opportunistic pathogen, since it remains benign until provided with specific conditions that allow it to cause disease. S. epidermidis was actually not considered a serious threat to human health prior to the introduction of catheters and surgery. Today, researchers and manufacturers are developing new approaches to designing catheters that prevent biofilm formation.
A Bacterial Sneeze, the Nose
The human nose is home to the infamous Gram positive bacterium Staphylococcus aureus, best known for its role in hospitals where it is a major cause of surgical wound and systemic infection. You may have heard of S. aureus in the media where it is often referred to as MRSA, standing for Methicillin Resistant Staphylococcus aureus. Infections of this bacterium are now a very serious threat to human health because it has become resistant to all commercially available antibiotics, including methicillin and vancomycin. It is often carried in the noses of health care workers and transmitted from patient to patient. Why some people carry S. aureus while others do not, is unknown.
A Mouthwash Away
It’s estimated that 500-600 different kinds of bacteria thrive on mucus and food remnants in the mouth. A predominant member of this community is the Gram positive bacterium Streptococcus mutans. It grows on biofilms on the surface of teeth (plaque) where it consumes sugar and converts it to lactic acid. Lactic acid erodes the enamel on the surface of teeth, which leads to the formation of cavities. Interestingly, a group of researchers have developed a strategy to combat dental decay by using a genetically modified strain of bacteria that produces a toxin that specifically kills S. mutans [3]. The trick is that this genetically modified strain of bacteria will only survive in your mouth if you provide it with specific nutrients. Basically, you brush the new strain of bacteria onto your teeth and they produce a toxin that prevents the growth of S. mutans thereby reducing the production of lactic acid. To maintain the strain of bacteria in your mouth you provide the essential nutrient by swishing daily with a mouthwash—just remember to feed your bacteria!
Whether or not tooth decay is a disease serious enough to warrant the use of a new strain of genetically modified bacteria is debatable. The effects of altering the populations of bacteria in the mouth may have unpredictable consequences. For example, Streptococcus pneumoniae is a much more threatening bacteria that can colonize the mouth. It’s an opportunistic pathogen that resides in the mouth and throat awaiting an opportunity to infect the lungs when defense systems are low, such as following an infection with influenza (the flu). Under normal circumstances the growth of S. mutans out competes the growth of S. pneumoniae in the mouth. Would the removal of S. mutans from your oral microflora result in increased growth of S. pneumoniae, and hence an increased risk of contracting pneumonia?
Braving Stomach Acid
What kind of organism would live in a highly acidic (pH 1-2) environment like the stomach? Not surprising there aren’t many organisms that have adapted to life in this environment. One organism that has been discovered living in the human stomach is the Gram negative bacterium called Helicobacter pylori [4]. How can it survive? Well, it creates a less acidic microenvironment. The bacteria achieve this by burrowing into the stomach’s mucosal lining to a depth where the pH is essentially neutral. In addition, H. pylori produce an enzyme called urease to convert urea produced by the stomach into ammonia and carbon dioxide.
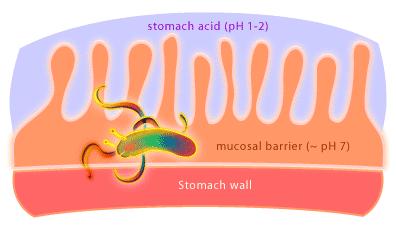
H. pylori is the causative agent of gastric ulcers, something once thought to be caused by stress, amongst other things, but which are now cured with antibiotics. Exactly how H. pylori causes ulcers is not yet known. It is thought that the bacterium may induce an immune response in the host that results in uncontrolled local inflammation and the generation of ulcers. Furthermore, it is now clear that gastric cancer is associated with H. pylori colonization as well. Research is underway to determine what role H. pylori is playing in this process. Overall, approximately 30-50% of the earth’s population is colonized by H. pylori and research has shown that H. pylori colonization varies from country to country and between socioeconomic and ethnic groups. How this transmission occurs is also unknown. Remarkably, ulcers develop in less than 20% of those people colonized by H. pylori. The fact that the majority of people colonized with H. pylori never develop ulcers has prompted microbiologists to suggest that H. pylori should be considered the normal flora of the human stomach. Some researchers believe that H. pylori may help to protect against conditions such as infant diarrhea and esophageal disease. Would the removal of H. pylori using antibiotics have unforeseen consequences for our inner ecosystem, and so our health? What exactly H. pylori is up to in the human stomach awaits further research.
Small Intestine vs. the Colon
Compared to the stomach, the small intestine is a relatively hospitable environment [5]. However, the small intestine presents microbes with a new challenge—high flow rates. This makes it difficult for bacteria to colonize the small intestine because they get washed out very quickly. As a result the concentration of bacteria in the small intestine remains relatively low (106 bacteria per ml) and human enzymes carry out most of the digestion processes. Minimizing the concentration of bacteria in the small intestine may be a strategy that our bodies have adapted in order to avoid microbial competition for high value nutrients such as simple sugars and proteins.
In the colon, things slow down. While it takes about 3-5 hours for food to move through the small intestine, it takes 24-48 hours for food to travel through the colon. This slower flow rate gives bacteria in the colon time to reproduce so that they reach very high concentrations (1012-1013 bacteria per ml). Bacteria packed into the lumen account for about 35-50% of the colon contents and for around 2 lbs of total body weight in an adult. The colon is a holding tank for bacteria that participate in the end stages of food digestion. For it is here that bacteria are presented with polysaccharides that cannot be broken down by human enzymes. The process of polysaccharide degradation in the colon is referred to as colonic fermentation. These polysaccharides are derived from plant material (eg. cellulose, xylan and pectin) and from human cells (eg. the polysaccharides that glue intestinal cells together) and are readily degraded by colonic bacteria. Polysaccharide fermentation results in the production of acetate, butyrate and propionate, which are used as a source of carbon and energy by mucosal cells of the colon. Thus, the colon can be considered an organ of digestion where bacteria do the majority of the work.
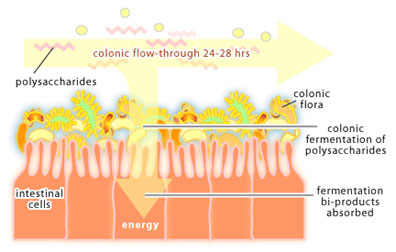
In the developed world, where nutrients are plentiful, colonic fermentation is not essential for survival. However, in areas where diets are high in plant polysaccharides and easily digestible nutrients are scarce, colonic fermentation could mean the difference between life and death. There is also evidence that E. coli within the colon produce vitamin K, which the human body requires for the process of blood clotting. The colon is a very complex microbial environment that we are only beginning to understand.
Vaginal
Relative to other microbial populations of the human body little is known about the normal flora of the vaginal tract. The predominant bacterial species are Lactobacillus. As is the case in other areas of the body, the presence of normal flora in the vaginal tract appears to have a protective role since women taking antibiotics for acne or urinary tract infections who have reduced levels of Lactobacillus often develop yeast infections. It is thought that Lactobacillus may prevent the growth of yeast by producing hydrogen peroxide, a bi-product of bacterial metabolism.
Bringing it All Together
The examples presented above describe a few examples of normal flora around the human body. From these examples several common themes can be extracted and to summarize, let’s discuss these themes:
1. Bacteria perform physiological, nutritional and protective functions in the human body.
2. Maintaining a balance is crucial. Normal flora consists of communities of bacteria that function as microbial ecosystems. If these ecosystems are disrupted the consequences can be unpredictable. Antibiotics, tissue damage, medical procedures, changes in diet, and the introduction of new pathogens are examples of changes that can affect your normal flora.
3. We are only beginning to appreciate the complexity and function of normal flora in the human body. Our understanding of microbial communities has been limited by our ability to culture microbes in the laboratory environment. It is thought that less that less than 1% of bacteria will grow on standard laboratory media. That means that we have yet to explore greater than 99% the microbial world. Today, new technologies such as the polymerase chain reaction (PCR), high-throughput DNA sequencing and DNA microarrays are starting to provide glimpses into these microbial ecosystems. Researchers have suggested that it is now time to embark on a “second human genome project [6]” where the genomic sequences of the microbes making up our normal flora are determined. Advancing our understanding of normal flora will provide us with fundamental information about who we are.
Additional Reading
1. Staley JT, Reysenbach AL, eds. 2002. Biodiversity of Microbial Life: Foundation of Earth’s Biosphere. New York: Wiley. 552p.
2. English MP. 1982. Microbes, Man, and Animals: The Natural History of Microbial Interactions. New York: Wiley. 342p.
3. Postgate JR. 2000. Microbes and Man. Oxford, UK; New York: Cambridge University Press. 373p.
4. Guarner F, Malagelada JR. 2003. Gut flora in health and disease. Lancet 361(9356): 512-9.
References
1. Salyers AA, Whitt DD. 2000. Microbiology: Diversity, Disease and the Environment. Bethesda Maryland: Fitzgerald Science Press.
2. Todar K. 2002. The Normal Bacterial Flora of Animals. University of Wisconsin Department of Bacteriology. Website
3. Hillman JD. 2002. Genetically modified Streptococcus mutans for the prevention of dental caries. Antonie Van Leeuwenhoek 82(1-4): 361-6.
4. Lynch NA. Helicobacter pylori and Ulcers: a Paradigm Revised. Federation of American Society for Experimental Biology. Website
5. Whitfield J. 2003. Gut reaction. Nature 423: 583-584.
6. Relman DA, Falkow S. 2001. The meaning and impact of the human genome sequence for microbiology. Trends Microbiol 9(5): 206-8.
(Art by Jiang Long)