RESTRICTION ENDONUCLEASES: MOLECULAR SCISSORS FOR SPECIFICALLY CUTTING DNA
(August 2003)
Today, in the age of molecular biology, the study of an organism’s genome (its complete DNA) is a central component driving our understanding of biology. When scientists first considered studying genomes they were faced with a problem: how to reproducibly cut a genome’s DNA into fragments that were small enough to handle? It was a significant problem. Genomes are composed of large DNA chunks on the order of millions of units, while a scientist could reasonable only handle pieces of DNA a few thousand units long. A discrepancy far too large to bridge, thus a method for reproducibly cutting DNA into manageable pieces was required to move the genomic studies forward. When this question was first posed in the 1970s it was becoming a relatively simple exercise to isolate DNA and then randomly cut it up using chemical or mechanical means. Unfortunately, this random cutting was not a satisfactory way to obtain smaller pieces of DNA, since it was impossible to tell what the original order of the DNA fragments were, an important point since the specific order of DNA is essential for its function. The biologists were stuck. A breakthrough was needed.
As with many of the important discoveries in biology, it was the study of bacteria that yielded this breakthrough. It was discovered that a type of bacterial enzyme was found to have the ability to cut DNA in a test tube [1-2]. These restriction endonucleases, so named because they cut double stranded DNA at restricted sites, were discovered as a natural part of the bacterial machinery. In a bacterial cell, restriction endonucleases (often referred to as restriction enzymes) act as a kind of immune system, protecting the cell from the invasion of foreign DNA, as would occur when a virus attempted to infect a bacterial cell. These restriction endonucleases provided biologists with a tool to study and manipulate DNA by enabling the generation of consistently sized DNA fragments. They are now used for a wide range of applications, including cloning, Southern hybridization analysis, DNA sequencing and global gene expression analysis (SAGE). Their importance to biotechnology is apparent when looking through the catalogue of any company that supplies reagents for molecular biological work-there are pages and pages devoted to listing different restriction enzymes. Lists that continue to grow as more enzymes are discovered. Truth be told, many recombinant DNA technologies, which the field of biotechnology heavily relies on, are unlikely to have been developed without the discovery of restriction enzymes.
The Discovery of Restriction Enzymes
Restriction endonucleases were discovered during experiments to determine the ability of a bacteriophage (the name given to viruses that infect bacteria) to infect two different laboratory strains of Escherichia coli called strain B and strain K [2]. In these experiments, bacteriophages were incubated with the different strains of E. coli and the ability of the bacteriophage to kill the E. coli cells was monitored. A bacteriophage will normally cause an infected bacterial cell to break open, killing the cell, while releasing millions of bacteriophage that had replicated within the cell. When bacteriophage spilling out of E. coli strain B cells were isolated, they were found to be very successful at re-infecting E. coli strain B cells. But when these bacteriophages were incubated with E. coli strain K, it was found that only a few of the bacteriophage could manage to replicate. This was a curious observation. Even more curious was the fact that after a series of incubations of the bacteriophage with strain K, the ability of the bacteriophage to infect and kill strain K increased. With a switch back to strain B, these bacteriophage that were now able to infect strain K to a high degree, showed a reduction of their ability to infect strain B (see Figure 1). It appeared that these bacteriophages, which were previously able to infect strain B to a high degree, had switched to become strain K-infective over the course of growing within strain K cells.
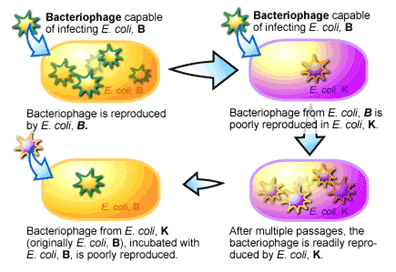
For the researchers, the results of this experiment suggested two things. The first was that strain K and strain B each had some mechanism that “restricted” the infective ability of bacteriophage isolated from the opposite strain. This was supported by the reduction of infectivity of the bacteriophage when it was initially switched from one strain to the other. The second was that the restriction of the bacteriophage’s infectivity was lost as the bacteriophage’s DNA was replicated inside the bacterial cell. This was evidenced by the return of the bacteriophage’s infectivity after several passages inside of the new host strain of bacteria.
What was this restrictive mechanism? It turns out that a bacterium “labels” its own DNA by modifying it in special ways. For example, in some bacteria the cytosine nucleotide has an extra single carbon group (referred to as a methyl group) added to it. This modification is referred to as methylation [3]. Each time a bacterium replicates its DNA these modifications are added to the newly synthesized DNA. When DNA comes from outside of the cell, as when a bacteriophage injects its DNA into a bacterium, it doesn’t have these modifications and thus is recognized as a foreign object to the cell. As you can imagine, a bacterium doesn’t want foreign and potentially harmful DNA floating around inside it and accordingly makes every attempt to degrade it when detected. To perform this function a series of enzymes evolved to chop up DNA that was not methylated-enzymes that were later named restriction endonucleases [2]. The existence of these enzymes explains the differences in the infection rate between E. coli B and K strains above. If bacteriophage DNA did manage to escape destruction, the replication of this foreign DNA by the bacterium over time would mark the bacteriophage DNA with the same modifications as the host, it would then no longer be recognized as foreign and be able to infect a bacterium at a much greater frequency. Discovery of this relatively simple system for detecting the difference between host and foreign DNA provided biologist with a new molecular tool: bacterial enzymes that could cut DNA, just like scissors cut paper.
How Restriction Enzymes Work and are Used
It has already been mentioned that restriction endonucleases recognize foreign DNA by their lack of methylation3, but the way these enzymes cut DNA is varied. Some cut the foreign DNA randomly. While others recognize a particular DNA recognition sequence and then either cut within this DNA sequence or several nucleotides away from it. Where a restriction endonuclease cuts within a DNA molecule is one of the primary characteristics by which it is classified [2]. For example, type II restriction endonucleases recognize a particular sequence and cut inside that sequence, while type III restriction endonucleases cut outside of their recognition site, the cut site being potentially a dozen nucleotide base pairs away or several thousand bases away.
Type II restriction endonucleases in particular have become an exceptional tool for the molecular biologist to specifically cut DNA. These enzymes bind to DNA at any position and then travel along the strand of DNA until they reach a recognition sequence [2]. Tight binding of the enzyme at the recognition site causes its structure to change, bringing the parts of the enzyme necessary for DNA cleavage into close proximity of the DNA strand. Once accomplished the “backbone” of the DNA molecule can be cut to produce two DNA fragments from one (see Figure 2). It is the recognition sites that provide the key to how one DNA fragment can be cut into two in a specific manner. These sites can vary in size; with some restriction endonucleases recognizing sequence motifs in DNA that are four nucleotides long, while others recognizing sequences that are twenty nucleotides long. The nucleotide pattern that is recognized by different restriction enzymes is quite variable, although it is frequently palindromic. A palindromic sequence is the same when read in 5′ to 3′ direction on either complementary strand of DNA, an example being the palindromic sequence recognized by the restriction enzyme known as EcoRI (see Figure 2).
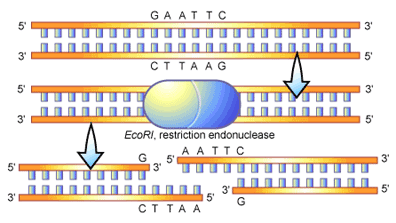
EcoRI recognizes a six-nucleotide pattern that reads GAATTC from the 5′ to the 3′ end of the DNA molecule. The complement of this sequence (on opposite DNA strand) also reads GAATTC when read from 5′ to 3′. In figure 2 you can see an illustration of an EcoRI molecule binding to and cleaving a strand of DNA, here you can see the palindromic recognition for the molecule. In this scenario the cleavage of the DNA molecule by EcoRI is symmetrical-with the enzyme cutting at the same point inside the sequence (between the G and the A when reading 5′ to 3′) on both strands. An important feature of this cleavage is the “overhanging” ends of the DNA molecule that are produced. These overhangs are often referred to as “sticky ends, ” since they can bind, or stick, to a complementary sequence of DNA (in this case 5′-AATT-3′). This stickiness is often utilized in the process of DNA cloning to help the adhesion of two DNA fragments. Overall, restriction endonucleases are quite variable in the DNA ends that they produce upon cleavage, for example leaving overhangs at the 5′ (as in EcoRI in figure 2), a 3′ overhang, or no overhang at all (referred to as “blunt” ends).
The frequency with which a given restriction endonucleases cuts DNA depends on the recognition site of the enzyme. As mentioned before, some enzymes recognize sites that are four nucleotides long (simply referred to as “four cutters”). With a quick calculation and a couple of basic assumptions, one can use this knowledge to estimate how frequently it should cut a piece of DNA. For example, with the four nucleotide bases that make up DNA the probability of any one nucleotide occurring at a given location is ¼. In the case of a “four cutter” a specific sequence of four nucleotides must be present and assuming that each nucleotide has an equal chance (i.e. ¼) of occurring at any particular site, then ¼ x ¼ x ¼ x ¼ = 1/256, a four-cutter should on average cut once every 256 base pairs.
A similar calculation can be applied to any restriction enzyme as long as the size of its recognition site is known, making it possible to predict the size and number of a DNA fragments that would be obtained by cutting a DNA molecule of known size. This fact gave molecular biologists the method they required to produce DNA fragments of known size for their experiments, as in gene mapping and genetic engineering.
Conclusion
Any catalogue of molecular biological reagents provides a sample of the great variety of restriction endonucleases available for research, and their names are quite strange: EcoRI, BamHI, BglII (pronounced “Bagel two”) and the racy SexAI. Their discovery has pushed the frontiers of molecular biology to greater things, allowing the development of techniques to create genetically modified proteins, to localize genes and to detect mutations that cause disease. Who would have thought that a pair of very small scissors could be so handy?
Text Consulted and Additional Reading
1. Old R.W., Primrose S.B., eds. (1994). Principles of Gene Manipulation: An Introduction to Genetic Engineering, 5th Ed. Blackwell Scientific Publications, U.S.A.
2. Burrell M.M., ed. (1993). Enzymes of Molecular Biology. Humana Press Inc., New York.
3. Kendrew J., ed. (1994). The Encyclopedia of Molecular Biology. Blackwell Scientific Publications, Oxford.
References
1. Meselson M., Yuan R. (1968). DNA restriction enzyme from E. coli. Nature 217: 1110-4.
2. Wilson G.G., Murray N.E. (1991). Restriction and Modification Systems. Annu. Rev. Genet. 25: 585-627.
3. Kessler C., et. al. (1985). Recognition sequences of restriction endonucleases and methylases—a review. Gene 33: 1-102.
(Art by Jane Wang – note that high res versions of image files available here)