STUDYING GENE FUNCTION: CREATING KNOCKOUT MICE
(August 2004)
Construction a knockout mouse
For decades researchers have tried to create tools that allowed for precise control over a specific gene in order to study its function. In the early 1980’s a breakthrough technology known as transgenics or gene transfer was developed [1]. This new technology involved the process of pronuclear microinjection, a method involving the injection of genetic material into the nuclei of fertilized eggs (Figure 1). Following injection, DNA would incorporate into the genome of the cell. The transformed fertilized eggs were then injected back into pregnant females and brought to term. A major downfall of this technique is that researchers could neither predict nor control where in the genome the foreign genetic material would be inserted. Since a gene’s location in the genome is important for its expression pattern, mouse lines carrying the same transgene could display wildly varying phenotypes.
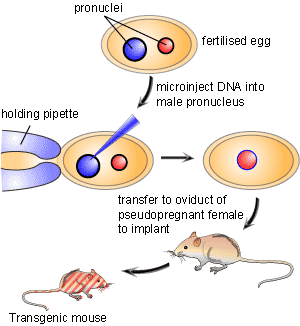
An innovative solution that resolved this conflict was originated by a team of scientists led by Martin Evans, Oliver Smithies and Mario Capecchi who created what is known as a genetic “knockout”. By creating a knockout they proved that it was possible to aim the inserted gene at a precise location in the mouse genome. This gave scientists the ability to replace, or knock out, a specific gene with an inactive or mutated allele. Therefore, knockout animals are considered an investigative technique that allows for a particular gene of interest to be removed in an attempt to define what effect that gene has in the life of the organism. The gene knockout is created by selectively disabling a specific target gene in a particular type of cells called embryonic stem cells. The three scientists received the Lasker Award in 2001 for this achievement. The technique has been used to make several thousand different knockout mice. Knockout mice have become one of the most useful scientific tools in helping to understand the human genome and its roles in disease.
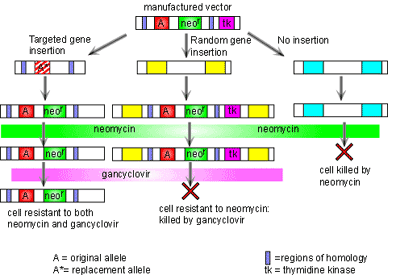
The process of gene targeting involves two processes. First a vector must be constructed. Once this has been done, the vector is put into embryonic stem cells and a new mouse is made from these cells [1]. The vector consists of two regions of homology at either end, the disrupted gene of interest, and two additional genes to allow for quality control in the process. The regions of homology are located at either end of the vector and are complementary to specific sites on the genome surrounding the gene of interest. The vector binds to the genome at these two points of homology. This technology then takes advantage of the process of recombination to swap the disrupted gene for the real one. Here the two additional genes are used. In order to make sure that the vector has been inserted into the genome, the neomycin resistance gene is included. If the vector has not been inserted then all cells will die when the cells are grown in neomycin. It is also possible that the vector has been inserted into the genome in the wrong place. To make sure this has not happened, the thymidine kinase (tk) gene is also added into the construct outside the region of homology. If recombination has taken place then the tk gene will not be in the genome and so cells will be insensitive to gancyclovir. However if the vector has been inserted into the genome completely then the cell will die when grown in gancyclovir. Thus through positive and negative selection, cells with the correct insert can be isolated. Figure 2 illustrates this process.
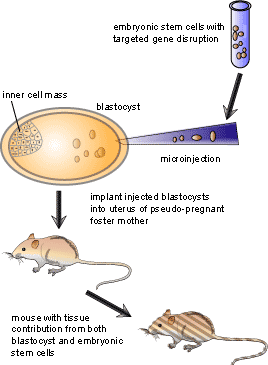
Embryonic stem cells are used as the target cells for the insertion of these vectors because they are pluripotent, meaning that they can give rise to every different type of cell in the adult body. Scientists interested in examining a specific gene will remove or “knock out” the gene in an embryonic stem cell as described above, then put the cell into a recently fertilized embryo. The manipulated cell then divides and eventually develops into many different tissues. The result is a mouse in which some of the cells have developed from the knocked-out embryonic stem cells. Often, embryonic stem cells which give rise to mice with a brown coat colour are injected into blastocysts which give rise to mice with a white coat colour. This allows the researcher to see which mice have contributions from both the injected cells and the blastocyst: if a mouse is completely white, then no injected cells grafted with the blastocyst. On the other hand if the mouse has both brown and white markings on its coat, then the injection of knockout embryonic stem cells was successful. After finding mice which have contributions from both cell types, the mice are bred to propagate the knockout gene to a certain percentage of the progeny, a process known as germ-line transmission. Once germ-line transmission is achieved, it is possible to breed mice where some are missing the knocked-out gene while others are not. In this case the latter group can act as a control group to see if the experiment has worked. Figure 3 illustrates the process of making knockout mice once the vector has been taken up by the cells.
Uses of knockout mice
Knockout mice are used in a variety of ways. They allow to test the specific functions of particular genes and to observe the processes that these particular genes could regulate. What actions does this gene turn off and on? By examining what is happening in an in vivo (inside the body) model, we are able to determine the effects a particular gene may have. These effects would be impossible to observe in a culture dish. However, to completely establish and assign an action to a particular gene is very challenging and requires an incredible amount of work.
One of the most exciting applications of knockout technology is in biomedical research. Scientists are using these models to study the progress of thousands of genetically based diseases at the molecular level. The hope is that by better understanding how a certain gene contributes to a particular disease, researchers can then take the knowledge a step further and look for drugs that act on that gene. Another essential application of knockout mice is in drug development. The knockout technology may lead to the discovery of the next generation of blockbuster therapies for curing numerous diseases based on novel targets from the human genome [4].
Although knockout technology is highly advantageous for both biomedical research and drug development, it also contains a number of limitations. For example, because of developmental defects, many knockout mice die while they are still embryos before the researcher has a chance to use the model for experimentation. Even if a mouse survives, several mouse models have somewhat different physical and physiological (or phenotypic) traits than their human counterparts. An example of this phenomenon is the p53 knockout. Gene p53 has been implicated in as many as half of all human cancers. However p53 knockout mice develop a completely different range of tumors than do humans. In particular, mice develop lymphomas and sarcomas, whereas humans tend to develop epithelial cell-derived cancers [2]. Because such differences exist it cannot be assumed that a particular gene will exhibit identical function in both mouse and human, and thus limits the utility of knockout mice as models of human disease.
More advanced types of knockouts
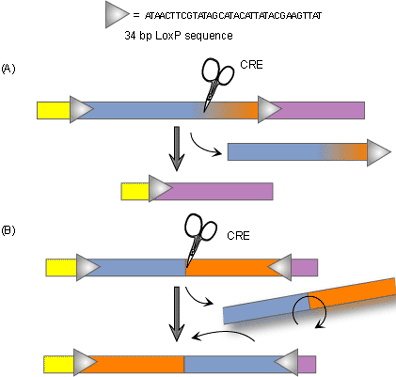
Recently, newer technologies have been developed which allow for conditional knockouts or tissue-specific gene targeting. The goal of conventional knockout technology is to knock out both alleles so that the gene is entirely absent from all cells. The purpose of conditional knockouts, in contrast, is to delete a gene in a particular organ, cell type, or stage of development [3]. Researchers can use the technique to knock out certain portions of specific genes at particular times when those genes are important. Conditional knockout mice have several benefits over the conventional type. Not only do they typically survive longer than traditional knockout mice, but conditional knockout methods are more precise as well. There are several different ways to make conditional knockout models, however the most widely used method is the Cre-loxP recombinase system. Cre recombinase is an enzyme that works like scissors to cut out a gene that is in between two target sequences called loxP. Because this enzyme is expressed only in certain cell types, the targeted gene will be knocked out of only those cells and only when the researcher wants them to be.
Disadvantages of knockout technology
Although knockout mice are extremely useful in studying gene function, producing custom knockout mice is very expensive. Typical cost ranges anywhere from $3,000 to as much as $30,000. There are hundreds of knockout mice commercially available. These can be purchased from a variety of companies, one being The Jackson Laboratory (Jax) in Bar Harbor, Maine.
It is also important to consider the cost of care for genetically altered animals, as they require special transgenic core facilities where staff can assist researchers with housing and taking proper measures to keep these knockout animals at optimal conditions for research purposes. The cost of equipping and maintaining such a facility is usually very high and may be a limiting factor.
Conclusions
In conclusion, knockout mice are a wonderful tool used to study the function of specific genes in a system. Despite numerous concerns and limitations applying this technique, it provides tremendous insight into understanding the disease process. The development of a gene knockout mouse has been a massive advance to the biomedical and pharmaceutical field presenting researchers with a very powerful tool for analyzing gene function during development, as well as in disease. In short, gene knockout technologies have become invaluable experimental tools for modeling genetic disorders, assigning functions to genes, evaluating drugs and toxins, and for helping to answer fundamental questions in basic and applied research. On top of it all, with constant evolution in biotechnology the future of knockout mice looks even friendlier!
References
1. (2002). The life history of the mouse in genetics. Nature 420: 510.
2. Pray L. (2002). Refining Transgenic Mice. The Scientist 16(13): 34.
3. Smith C.M. (2000). Technical Knockout. The Scientist 14(15): 32.
4. Zambrowicz B.P., Sands A.T. (2003). Knockouts Model the 100 Best-Selling Drugs—Will They Model the Next 100? Nature Reviews Drug Discovery 2: 38-51.
Additional Reading
1. Nagy A. (2000). Cre recombinase: The universal reagent for genome tailoring. Genesis 26(2): 99-109.
(Art by Fan Sozzi – note that some high res versions of these image files available here)