CLEAVAGE OF SUPERPOWER PROTEIN DETERMINES THE SUPERPOWER PHENOTYPE IN A TISSUE DEPENDENT MANNER: HOW INCREDIBLE!
JOURNAL OF SUPERHERO RESEARCH (2013). Vol 13 Vol 547. pp328-3332 doi:23.1724/SR20130328 pdf download
We demonstrated that the presence of superpower (SP) protein cleavage is responsible for the diversity of superpowers in a tissues-specific manner within The Incredibles family. This cleavage event produces two fragments, 135 kDa and 85 kDa, without a loss of peptide, as confirmed by Edman degradation. We speculate that the 85 kDa protein fragment is involved in the generation of superpower.
Key words: Superpower phenotype, Supers, Supers Relocation Program, The Incredibles.
INTRODUCTION
The study of humans with unusual intrinsic superpowers, or Supers, has been an ongoing effort in parallel to the government’s “Superhero Relocation Program”, with aims to integrate the Supers among the civilians and not use their abilities [1].
A group of investigators at the National Supers Agency has recently sequenced a novel gene, superpower (SP), within the Supers population [2]. Interestingly, this SP gene is also present in the non-Supers civilians, though with a drastically reduced expression [3]. Therefore, it is hypothesized that enhanced SP expression contributes to Supers’ diverse superpower phenotypes [3].
The Incredibles family is the first known Supers family, composed of the two parents – Bob and Helen Parr, as well as their three children- Violet, Dashielle (Dash), and Jack-Jack. Each family member exhibits a unique superpower: Mr. Parr possesses incredible strength, while Mrs. Parr is able to stretch any part of her body up to 34m in length and 1mm thin. Their children, however, display a wide range of superhero phenotypes that differ from the parents, such as invisibility, extreme high speed, and shape-shifting [4]. This characteristic made The Incredibles family ideal for investigating whether or not enhanced SP expression is responsible for their respective superpower phenotypes, while minimizing genetic variation between subjects. We hypothesize that the SP gene undergoes post- transcriptional or post-translational modification to yield varied superpowers in this family. The knowledge gained from this study will provide insights into how to devise molecular strategies to manage and to attenuate the superpowers of the Supers. Ultimately, the results of the study will contribute towards an overall increase in general public safety and will reduce the cost of national court service [5,6].
EXPERIMENTAL
Human Subjects and Tissues
The study was approved by the ethics committee of the University of Metroville and written informed consent was obtained from all members of The Incredibles family as well as age- and gender-matched non-Supers controls. Parental consent was obtained for participants who are under the age of 21. Buccal cell extraction was performed on all participants. Whole blood was obtained in parallel by venipuncture, using acid citrate dextrose (0.18M glucose, 0.08M trisodium citrate, 0.052M citric acid) as anticoagulant in a final ratio of 1:5 (v/v). Skeletal muscle of arms (bicep muscles) and legs (gastrocnemius muscles), connective tissue proper of dermis, and epithelial cells biopsies were extracted from all participants. Muscle biopsies were carried out via needle percutaneous biopsies.
DNA/RNA/Protein Extraction
The extraction was performed with the AllPrep DNA/RNA/Protein Mini Kit (Qiagen Inc, Valencia, CA, USA) according to a modified manufacturer’s protocols. For extraction from whole blood, 500μL of samples was disrupted by adding 500μL of Buffer RLT (AllPrep kit). For extraction from tissues and pelleted buccal cells, 10mg (approximately 1mm3) of each trypsinized cell type were used and disrupted by adding 350μL of Buffer RLT (AllPrep kit). The RLT buffer was supplemented with β-mercaptoethanol as detailed in the manufacturer’s instruction. For the tissues and whole blood, the homogenization was performed on ice with a blunt 20-gauge needle (0.9mm diameter) fitted to an RNase-free syringe. Further steps were carried out according to the AllPrep’s instruction manual. Following extractions, the absorbance at 260 and 280nm was measured using a NanoDrop ND-1000 spectrophotometer (Thermo Fisher Scientific, Wilmington, DE, USA), and the ratios were calculated.
DNA Sequencing of SP gene
The SP gene was amplified using the primers (forward: 5′-GCTCATAACGTCCTGACCAG-3, reverse: 5′-GTCGAACGTGATCGCTAGTC-3’) [2,3]. Each 25μL PCR mixture contained 12.5μL HotStarTaq master mix (Qiagen Inc, Valencia, CA, USA), 0.15μL of the forward and reverse 50μM primers, 7.2μL nuclease free water, and 5μL of genomic DNA. PCR was performed on a MyCycler (Bio-Rad, Hercules, CA, USA) and included an initial denaturation step at 95°C for 15 min, followed by 35 cycles of denaturation at 95°C for 30 sec, annealing at 60°C for 30 sec, and elongation at 72°C for 30 sec, with a final elongation step at 72°C for 7 min. PCR products were analyzed on 1% agarose-gels. Electrophoresis was carried out at 80V for 1h. Verified PCR products were purified using MinElute® PCR Purification Kit (Qiagen Inc, Valencia, CA, USA). Purified PCR products were measured spectrophotometrically, diluted with nuclease free water, mixed with primers then submitted to GenePixar (GenePixar Inc; Emeryville, CA, USA) for DNA sequencing and uploaded to GenBank (BB041027-BB050315).
Real-time RT-PCR analysis of SP gene
Expression of SP mRNAs were determined by real-time RT-PCR using SYBR Green fluorescence on a LightCycler instrument (Roche Diagnosis; Manheim, Germany) using the standard curve method. The constitutively expressed β-actin was used to verify uniform sample loading. The PCR amplification reactions and preparation of standards were performed as previously described [3]. Primers for real-time RT-PCR were: SP forward: 5’-  ; reverse: 5’-  ; β-actin forward: 5’-AGGCACCAGGGCGTGAT-3’, reverse: 5’-GCCCA CATAGGAATCCTTCTGAC -3’. The qPCR was done at 95°C for 10 min followed by 40 cycles at 95°C for 15 s and 60°C for 30 s. The specificity of PCR product was examined by dissociation curve, and results calculated by the 2-ΔΔCT method.
Western Blot
Protein concentration was measured using the BCA protein assay kit (Bio-Rad; Hercules, CA, USA). After denaturation at 95°C, proteins were separated by 4-20% SDS-PAGE. Separated proteins were transferred onto polyvinylidene fluoride (PVDF) membranes, which were washed in TBST before incubation in 5% skim milk (diluted in TBST) at 37°C for 1 h. Blots were incubated overnight with polyclonal rabbit anti-human SP antibody (Sigma, Saint Louis, MO, USA) at 37°C, and then incubated with secondary goat anti-rabbit IgG antibody (Sigma, Saint Louis, MO, USA) at 37°C for 1 h. Immunoreactive bands were detected by enhanced chemiluminescence. Bands were analyzed using ImageJ software (version 1.44p, USA). Values obtained were normalized basing on density values of internal β-actin.
Protein Sequencing of SP Fragment S
The stained protein bands were excised from the PVDF membrane and directly applied to the upper glass block of the reaction chamber in Procise 494 gas-phase protein sequencer (Applied Biosystems, Foster City, CA, USA). Edman degradation is performed according to the standard program supplied by Applied Biosystems. The released phenylthio-hydantoin (PTH) amino acids are separated by an online high-performance liquid chromatography system and identified by retention time.
Statistical Analysis
mRNA expression levels between The Incredibles family and the non-Supers controls were made by paired t- test. Two-way ANOVA was used to compare the mean protein expression levels between different cell types of the two groups. This was followed by Tukey’s post-hoc test. A p-value of 0.05 was considered statistically significant.
RESULTS AND DISCUSSION
The SP gene has been identified in both the Supers and non-Supers, with differential gene expression levels [2,3]. In an attempt to decipher the genetic influence SP expression has on the diverse superpower phenotypes in the Supers, we focused on the SP gene expression in members of The Incredibles family. They were chosen as our study subjects to minimize genetic background variation and because each family member exhibits different superpowers.
It was crucial to confirm that the distinct superpower phenotypes observed within the family was not a consequence of somatic mutation in the SP DNA sequence (data not shown). Identical SP DNA sequence between all family members suggests that phenotypic differences may be a result of differences in SP protein expression.
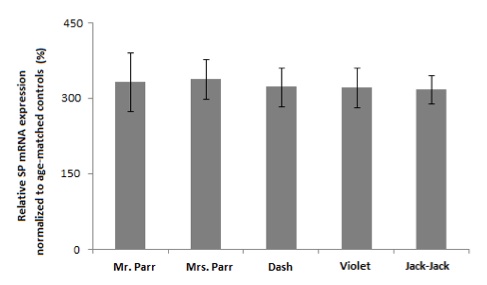
We first examined the SP mRNA expression using readily accessible buccal cells as our specimen (Figure 1). Assuming that SP indeed plays a role in superpower phenotypic manifestation, similar mRNA expression level between the the Non-Supers and The Incredibles (p > 0.05) implies that the effect of SP on superpower phenotypes is not due to transcriptional differences. The observed discrepancy in phenotype may be explained by post-translational modification. Whole blood samples yielded similar mRNA expression levels between the two groups (data not shown).
Figure 1. [Click Image to Enlarge] SP protein is up-regulated in buccal cells of Violet and Jack-Jack. Graph represents relative SP mRNA expression normalized to actin. Error bars denote ±SEM. N=5 per group. (***P<0.001 vs age-matched control; ###P< 0.001 vs. family member counterparts.)
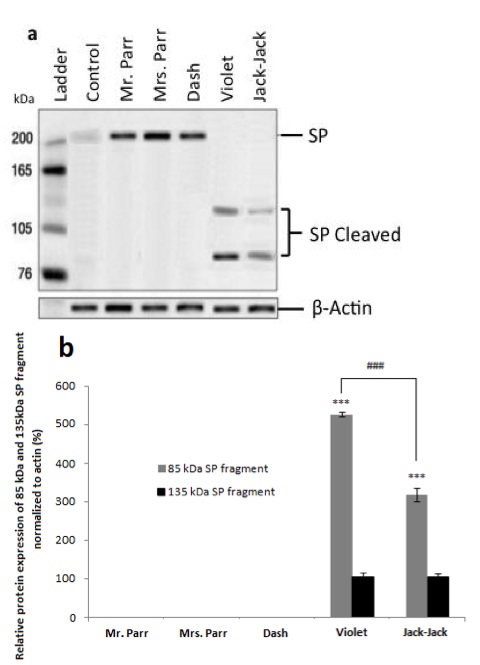
In accordance with previous findings [3], the non-Supers control exhibits lowlevels of SP protein (Figure 2). Differential protein expression levels were observed in buccal cells between the family members using polyclonal rabbit anti-SP antibody (Figure 2). Dash, Mr. and Mrs. Parr have identical protein bands as that of non-Supers control at 220 kDa, while Violet and Jack-Jack show two protein bands at 135 kDa and 85 kDa (Figure 2).
Figure 2. [Click Image to Enlarge] SP protein expression in buccal cells. Western blot analysis of SP protein a) and relative SP protein expression b) of The Incredibles family, normalized to actin . Error bars denote ±SEM. (N=5 per group; P
Since we do not observe superpowers in civilians, it is plausible that the uncleaved 220kDa SP protein does not contribute to superpower phenotypes. Following the same logic, this may indicate that Dash, Mr. and Mrs. Parr’s superpowers do not involve epithelial cells. Violet’s invisibility superpower is due to her ability to manipulate her epithelial pigmentation [4]. Since the cleaved form of SP is present only in Violet’s and Jack-Jack’s buccal cells, it leads us to speculate that the cleaved SP fragments are correlated with the presence of superpower. However, the cleaved SP fragments in Jack-Jack’s buccal cells show a reduced expression level compared to that of Violet’s (Figure 2b). It suggests that while shape-shifting may require the manipulation of epithelial cells, it is to a less degree than invisibility. We did not observe any differential protein expression in whole blood (data not shown), which may signify that circulatory system is not directly involved in The Incredibles’ superpowers. These data allude to tissue-specific SP protein cleavage, which relate to the various display of superpowers apparent in the family. To confirm our inference, we obtained skeletal muscles from gastrocnemius of the calf and bicep of the upper arm, as well as connective tissue of dermis from all subjects.
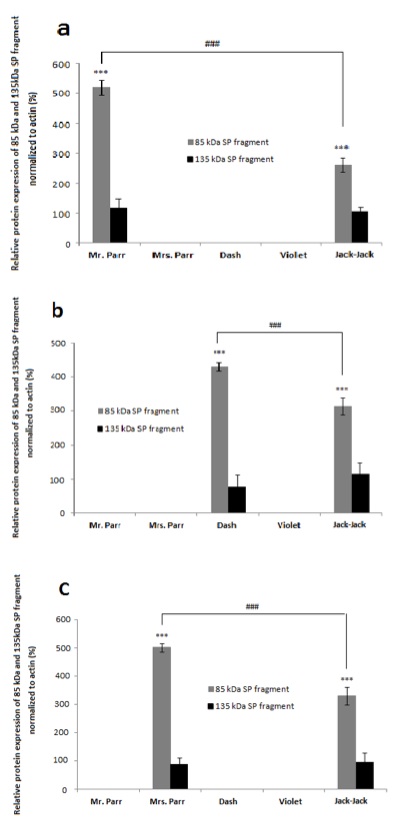
We detected SP protein cleavage in specific tissues for each family member: in bicep muscle of Mr. Parr (Figure 3a), gastrocnemius muscle of Dash (Figure 3b), and connective tissue of Mrs. Parr (Figure 3c). This observation provides support for our speculation that cleaved SP fragments are correlated with superpower display, as the presence of cleaved protein products coincide with Mr. Parr’s enormous upper body strength, Mrs. Parr’s tremendous stretchability, and Dash’s lightning speed [4]. Though Jack-Jack has yet to fully develop his defined superpower phenotype, he seems to demonstrate the most versatile abilities in the family [4]. This is also reflected in the protein expression, whereby the SP cleavage event is present in all tissues observed, unlike the rest of the family members with SP cleavage confined to a single tissue (Figure 3). Taken together, the cleaved SP protein seems to determine the superpower phenotype in a tissue-specific manner.
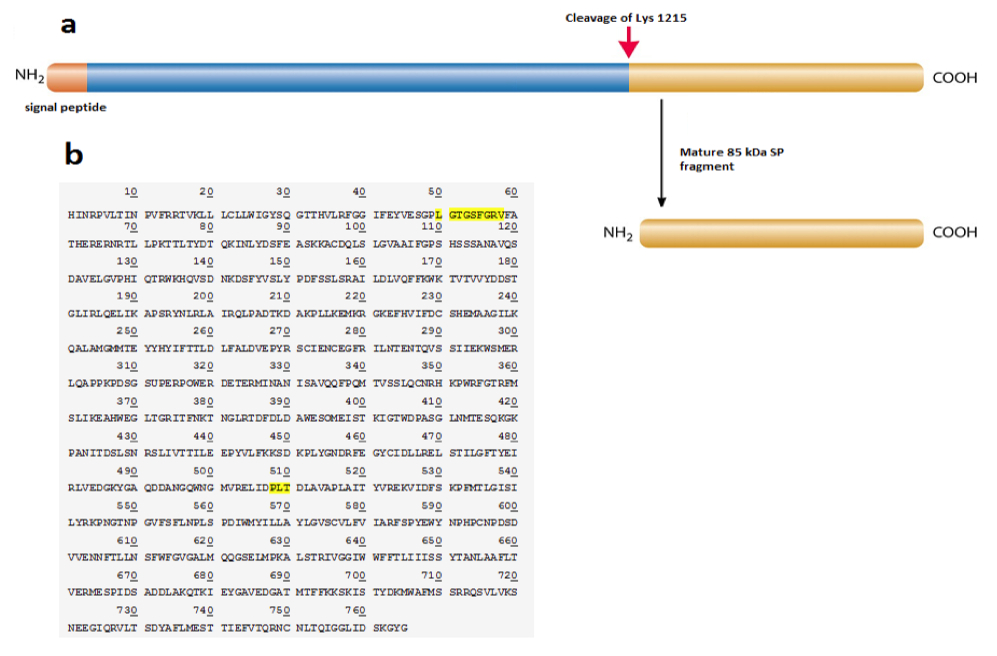
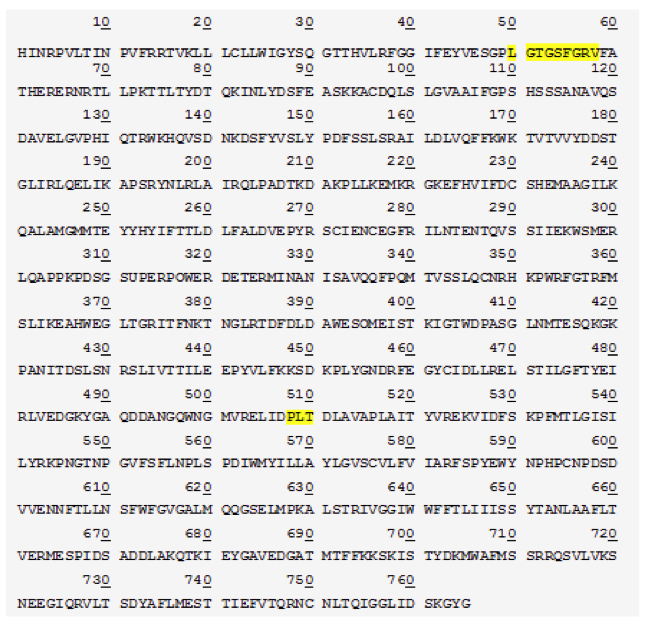
Figure 3. [Click Image to Enlarge] SP protein is cleaved at Lys 1215 to yield 85 kDa fragment. a) Schematic of the SP protein showing the cleavage site and products, b) protein sequence of the 85 kDa SP fragment by Edman sequencing. Regions highlighted represent functional domains (ATP-binding site at position 50-58; glutamate binding site at position 508-510).
In an event of SP protein cleavage, there is a significant increase in the 85kDa SP fragment expression compared to that of the 135 kDa fragment (Figure 2). This led us to reason that perhaps the 85kDa fragment is the functional determinant and contributes to the displayed superpower phenotype while the 135 kDa SP undergoes degradation or removal following tissue-specific cleavage. The two smaller protein fragments add up to 220 kDa, suggesting that there is a cleavage event of SP without loss of peptides (Figure 2). This is confirmed by protein sequencing of both the SP protein as well as the 85 kDa fragment. Results show that the SP protein is composed of 1980 amino acids. The cleavage site is at lysine 1215 which yields the 85 kDa fragment composed of 765 amino acids (Figure 4a). A candidate protease that performs this function may be a protease within the trypsin-like serine protease family, which is able to cleave peptide bonds following a positively charged amino acid such as lysine or arginine. We further studied the SP protein sequence with aims to understand its potential functionality (Figure 4b). Comparison with the Genbank protein database uncovered conserved protein sequences correlated with known, defined functions. Position 50-58 codes for an ATP- binding site, which implies that ATP may be required for appropriate fragment 85kDa func tion (Figure 4b). A glutamate binding site at position 508-510 was also identified and is found to be homologous to human glutamate receptor 3 protein sequence (Figure 4b).
Figure 4. [Click Image to Enlarge] The 85 kDa SP protein fragment exhibit tissue- specific expression among Incredibles family members with different super powers. Graphs represent relative SP protein expression in a) bicep muscles, b) gastrocnemius muscles, and c) the connective tissue proper of the dermis, normalized to actin.
Therefore, it is possible that glutamate is implicated in the signal transduction pathway of this peptide. This finding offers a potential explanation for the ability of the Supers to exert control over their powers, as it is possible that this peptide may interact with glutamate to modulate electrical signal transduction within peripheral neural network.
In this study, we have demonstrated that elevated SP mRNA and protein expression are correlated with individuals possessing superpowers. Upon further examination, it appears that the SP protein undergoes cleavage in a tissue-specific manner to yield 85 kDa and 135 kDa fragments, and the cleavage event is correlated with the tissue type that display the supernatural powers . Within The Incredibles family, we observe an elevated expression of the 85 kDa fragment while the 135 kDa fragment expression is reduced. We therefore propose that the 85 kDa fragment is the functional unit of the SP protein, and contribute to the molecular pathways necessary to generate superpowers. The result from this investigation is limited to The Incredibles family. The SP protein cleavage and its effect on the superpower phenotypes require further validation in a greater population of Supers. The physiological function of SP 85kDa fragment may be further examined in animal models, with aims to confirm the hypothesized SP contribution to superpower phenotype in a tissue-dependent manner and to elucidate the molecular and signaling pathways involved in manifestation of enhanced abilities.
Furthermore, it would be beneficial to study the physiology of full-length PS protein, the mode of inheritance of SP gene, and the regulation of SP cleavage, as it may allow us to optimally attenuate of superpower in potential Supers’ offspring and better integrate them into the harmonious non-Supers society.
ACKNOWLEDGEMENT
This work was conducted in collaboration with Dr. Rick Dicker at the Institute of Supers Relocation Program Research. We thank Dr. Edna Mode for her expertise and technical assistance in tissue biopsy procedures. We express our most sincere appreciation and gratitude for the subjects’ participation and cooperation throughout the study.
FUNDING
This work was made possible with support from the National Supers Agency under Grant 4- 2013-515.
REFERENCES
1. Superhero Relocation Program Act. 2000. National Supers Agency, US Government.
2. Dicker, R., Griffin, P., Pekoe. T. Novel gene Superpower sequenced in Supers of Westburg, California. J. Super Res. 2010. 35:184-190.
3. Lee, B., Chan, J., Lee A. Chinese Supers shows enhanced Superpower gene expression compared to non-Supers Chinese in Kongfu-ville. Intl. Genet. 2011. 23:782-790.
4. National Supers Agency Superpower Database of the United States. National Supers Agency, US Government.
5. Kitler A.,Massolino B.,Staller J. Evaluation of Super-Villains in International Supers Confinement Institution on public safety and national security. J. Def Tech. 2009. 8374: 53282-53305.
6. Supers Relocation Program Annual Report: Metroville 2012. National Supers Agency, US Government.
In case it wasn’t clear already, this article is not real.