WHAT IS BIOINFORMATICS?
Bioinformatics involves the integration of computers, software tools, and databases in an effort to address biological questions. Bioinformatics approaches are often used for major initiatives that generate large data sets. Two important large-scale activities that use bioinformatics are genomics and proteomics. Genomics refers to the analysis of genomes. A genome can be thought of as the complete set of DNA sequences that codes for the hereditary material that is passed on from generation to generation. These DNA sequences include all of the genes (the functional and physical unit of heredity passed from parent to offspring) and transcripts (the RNA copies that are the initial step in decoding the genetic information) included within the genome. Thus, genomics refers to the sequencing and analysis of all of these genomic entities, including genes and transcripts, in an organism. Proteomics, on the other hand, refers to the analysis of the complete set of proteins or proteome. In addition to genomics and proteomics, there are many more areas of biology where bioinformatics is being applied (i.e., metabolomics, transcriptomics). Each of these important areas in bioinformatics aims to understand complex biological systems.
Many scientists today refer to the next wave in bioinformatics as systems biology, an approach to tackle new and complex biological questions. Systems biology involves the integration of genomics, proteomics, and bioinformatics information to create a whole system view of a biological entity.
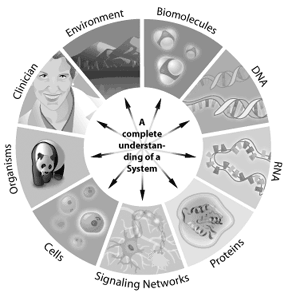
For instance, how a signaling pathway works in a cell can be addressed through systems biology. The genes involved in the pathway, how they interact, and how modifications change the outcomes downstream, can all be modeled using systems biology. Any system where the information can be represented digitally offers a potential application for bioinformatics. Thus bioinformatics can be applied from single cells to whole ecosystems. By understanding the complete “parts lists” in a genome, scientists are gaining a better understanding of complex biological systems. Understanding the interactions that occur between all of these parts in a genome or proteome represents the next level of complexity in the system. Through these approaches, bioinformatics has the potential to offer key insights into our understanding and modeling of how specific human diseases or healthy states manifest themselves.
The beginning of bioinformatics can be traced back to Margaret Dayhoff in 1968 and her collection of protein sequences known as the Atlas of Protein Sequence and Structure[1]. One of the early significant experiments in bioinformatics was the application of a sequence similarity searching program to the identification of the origins of a viral gene[2]. In this study, scientists used one of the first sequence similarity searching computer programs (called FASTP), to determine that the contents of v-sis, a cancer-causing viral sequence, were most similar to the well-characterized cellular PDGF gene. This surprising result provided important mechanistic insights for biologists working on how this viral sequence causes cancer[3]. From this first initial application of computers to biology, the field of bioinformatics has exploded. The growth of bioinformatics is parallel to the development of DNA sequencing technology. In the same way that the development of the microscope in the late 1600’s revolutionized biological sciences by allowing Anton Van Leeuwenhoek to look at cells for the first time, DNA sequencing technology has revolutionized the field of bioinformatics. The rapid growth of bioinformatics can be illustrated by the growth of DNA sequences contained in the public repository of nucleotide sequences called GenBank.
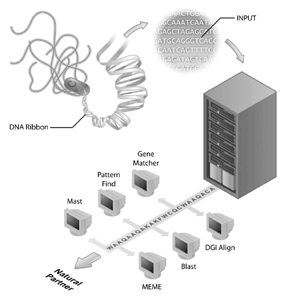
Genome sequencing projects have become the flagships of many bioinformatics initiatives. The human genome sequencing project is an example of a successful genome sequencing project but many other genomes have also been sequenced and are being sequenced. In fact, the first genomes to be sequenced were of viruses (i.e., the phage MS2) and bacteria, with the genome of Haemophilus influenzae Rd being the first genome of a free living organism to be deposited into the public sequence databanks[4]. This accomplishment was received with less fanfare than the completion of the human genome but it is becoming clear that the sequencing of other genomes is an important step for bioinformatics today. However, genome sequence by itself has limited information. To interpret genomic information, comparative analysis of sequences needs to be done and an important reagent for these analyses are the publicly accessible sequence databases. Without the databases of sequences (such as GenBank), in which biologists have captured information about their sequence of interest, much of the rich information obtained from genome sequencing projects would not be available.
The same way developments in microscopy foreshadowed discoveries in cell biology, new discoveries in information technology and molecular biology are foreshadowing discoveries in bioinformatics. In fact, an important part of the field of bioinformatics is the development of new technology that enables the science of bioinformatics to proceed at a very fast pace. On the computer side, the Internet, new software developments, new algorithms, and the development of computer cluster technology has enabled bioinformatics to make great leaps in terms of the amount of data which can be efficiently analyzed. On the laboratory side, new technologies and methods such as DNA sequencing, serial analysis of gene expression (SAGE), microarrays, and new mass spectrometry chemistries have developed at an equally blistering pace enabling scientists to produce data for analyses at an incredible rate. Bioinformatics provides both the platform technologies that enable scientists to deal with the large amounts of data produced through genomics and proteomics initiatives as well as the approach to interpret these data. In many ways, bioinformatics provides the tools for applying scientific method to large-scale data and should be seen as a scientific approach for asking many new and different types of biological questions.
The word bioinformatics has become a very popular “buzz” word in science. Many scientists find bioinformatics exciting because it holds the potential to dive into a whole new world of uncharted territory. Bioinformatics is a new science and a new way of thinking that could potentially lead to many relevant biological discoveries. Although technology enables bioinformatics, bioinformatics is still very much about biology. Biological questions drive all bioinformatics experiments. Important biological questions can be addressed by bioinformatics and include understanding the genotype-phenotype connection for human disease, understanding structure to function relationships for proteins, and understanding biological networks. Bioinformaticians often find that the reagents necessary to answer these interesting biological questions do not exist. Thus, a large part of a bioinformatician’s job is building tools and technologies as part of the process of asking the question. For many, bioinformatics is very popular because scientists can apply both their biology and computer skills to developing reagents for bioinformatics research. Many scientists are finding that bioinformatics is an exciting new territory of scientific questioning with great potential to benefit human health and society.
The future of bioinformatics is integration. For example, integration of a wide variety of data sources such as clinical and genomic data will allow us to use disease symptoms to predict genetic mutations and vice versa. The integration of GIS data, such as maps, weather systems, with crop health and genotype data, will allow us to predict successful outcomes of agriculture experiments. Another future area of research in bioinformatics is large-scale comparative genomics. For example, the development of tools that can do 10-way comparisons of genomes will push forward the discovery rate in this field of bioinformatics. Along these lines, the modeling and visualization of full networks of complex systems could be used in the future to predict how the system (or cell) reacts, to a drug, for example. A technical set of challenges faces bioinformatics and is being addressed by faster computers, technological advances in disk storage space, and increased bandwidth, but by far one of the biggest hurdles facing bioinformatics today, is the small number of researchers in the field. This is changing as bioinformatics moves to the forefront of research but this lag in expertise has lead to real gaps in the knowledge of bioinformatics in the research community. Finally, a key research question for the future of bioinformatics will be how to computationally compare complex biological observations, such as gene expression patterns and protein networks. Bioinformatics is about converting biological observations to a model that a computer will understand. This is a very challenging task since biology can be very complex. This problem of how to digitize phenotypic data such as behavior, electrocardiograms, and crop health into a computer readable form offers exciting challenges for future bioinformaticians.
(This article is based upon an interview with Francis Ouellette, Director of the UBC Bioinformatics Centre)
References
1. PMID=5789703; Sci. Am. 1969 Jul; 221(1):86-95.
2. PMID=6304883; Science. 1983 Jul 15; 221(4607):275-7.
3. PMID=6306471; Nature. 1983 Jul 7-13; 304(5921):35-9.
4. PMID=7542800; Science. 1995 Jul 28; 269(5223):496-512.
(Art by Jiang Long – note that high res versions of image files available here)