HOWLETT: NOVEL WOLVERINE PROTEIN CONTRIBUTES TO RAPID REGENERATION AND HEIGHTENED CELLULAR REPLICATION
JOURNAL OF SUPERHERO MUTATIONAL SCIENCE (February 2013). Vol 13 Issue 2. pp297-302 pdf download
Wolverine, a mutant from the X-men team, possesses super healing abilities. Wolverine’s healing abilities have striking similarities to that seen in axolotl, an amphibian with the capacity to regenerate amputated limbs. In this study we sought to determine the mechanisms by which Wolverine regenerates. We identified a novel protein, dubbed Howlett, that is nearly identical to the Amblox protein in axolotl which is known to be responsible for the amphibian’s limb regeneration. siRNA knockdown of the howlett and amblox genes demonstrated decreased replication in Wolverine and axolotl, respectively, and Howlett was found in all Wolverine tissues. Using mass spectrometry and x-ray crystallography, we identified an S2 pocket in the Howlett protein that we postulate contributes to the 5.66-fold increased specific activity observed over Amblox in cleaving a large substrate analogue. Our findings show that Howlett is a major contributor to Wolverine’s incredible regeneration capacity, and further investigation of the signaling and regulatory mechanisms associated with this novel protein could provide outstanding advances in the field of regenerative medicine.
Wolverine (aka Logan or James Howlett) is a member of the X-Men, a team of mutant superheroes. Wolverine possesses superhealing abilities, known as Healing Factor, which allows him to recover from any wound, disease or toxin faster than human rate. This Healing Factor permitted the bonding of adamantium, an indestructible metal alloy, to his skeleton when he was subjected to the Weapon X program, giving him his signature claws. Healing Factor also slows down the aging process in Wolverine, allowing him to live beyond the normal human lifespan.
Our group has been investigating the regeneration and wound healing capacity of the axolotl (Ambystoma mexicanum), a neotenic amphibian with the ability to regenerate amputated limbs1. Our research on the axolotl has implications in the field of Regenerative Medicine to help patients such as amputees, transplant recipients and burn victims. We and others have found the same mechanism that axolotl uses for regeneration and wound healing. This mechanism involves a protein called Amblox, which possesses protease functions and may be involved in the replication and dedifferentiation capacity of cells at the wound site. We have observed that the axolotl and Wolverine have striking similarities in their ability to regenerate. In this study, we sought to determine the mechanisms of Wolverine’s Healing Factor.
We have recently identified the amblox gene, and a collaborator has also generated a monoclonal antibody against the Amblox protein for use in Western Blot and ELISA analyses. Using PCR, we have identified a homologous gene in Wolverine, giving. In addition, using the α-Amblox mAb, we have identified a protein, Howlett, that is nearly identical to the Amblox protein found in the axolotl. Finally, we charaterized the efficiency of the protein to account for the increased speed of Wolverine’s healing abilities.
MATERIALS & METHODS
Animals. Axolotls (Ambystoma mexicanum) of 4-7 inches in length were purchased from the Ambystoma Genetic Stock Centre (University of Kentucky, Lexington, KY, USA). Animals were maintained in 40% Holtfreter’s solution and treated in accordance with Xavier University’s Animal Care Committee’s regulations. For all experiments, group sizes were between two to four animals.
Human and mutant tissues. Control tissue, from three normal humans and three X-Men mutants with different abilities, and Wolverine tissue for PCR and protein assays were obtained from epidermal scrapings from subjects. To test for presence of Howlett in other tissues, biopsies were taken from various Wolverine tissues, with ethical approval from the Xavier University Research Ethics Board.
Culture of axolotl and Wolverine cells. Epidermal cells harvested from axolotl and Wolverine were trypsinized and grown in DMEM with 10% FBS, 1% Pen/strep and 1% Glutamax. After 12 hours, cells were 80% confluent and passaged. Cell cultures were kept at 37oC with 5% CO2.
Polymerase chain reaction. PCR was performed on extracted DNA from biopsies and also from cultures using axolotl amblox specific primers and a Taqman PCR kit (Invitrogen) under the following conditions: 35 cycles of 95°C for 30s, 60°C for 30s and 72°C for 30s and an extension time of 72°C for 5 min. Our amblox primers were: Fwd 5’-agtcgtggctagtcatg ctgtagttcaatgaa-3’ and Rev 5’- gtacgatcggatagaacttccagctagt-3’, generating a 297bp fragment. The amplified genes were detected by electrophoresis in a 1.5% agarose gel with ethidium bromide (500ng/ml) and bands visualized using Gel Doc 2000 (Bio-Rad Laboratories)
siRNA Knockdown. Cells were plated onto six-well plates (106 per well); maintained in antibiotic-free medium for 24h; and transfected with a mixture containing Opti-MEM, 10μL/well Lipofectamine (Invitrogen), and either 0.5μg/well scramble siRNA or amblox siRNA (generated by Dr. Scott Summers) for 5h. The sequence of these siRNAs is the intellectual property of Dr. Summers and we are therefore not at liberty to release it. Cells were then incubated with fresh medium for 1h and were either lysed or used for BrdU incorporation assay.
BrdU incorporation assay. BrdU (Bromodeoxyuridine) DNA labeling was performed on 10 cells with 100 μM of BrdU for 2 hours to determine the percentage of dividing cells, and analyzed with anti- BrdU FITC (Bioscience) as per manufacturer’s instructions. The percentage of dividing cells was determined by flow cytometry.
Mass spectrometry. Mass spectrometry was performed by our collaborator Dr. Leonard Foster at the Centre for High-Throughput Biology at the University of British Columbia as previously described2.
Analysis of mass spectrometry data. See online Supplementary Methods.
Crystallization of Howlett and Structure Determination and Refinement. Crystals were obtained under various conditions, and optimization was carried out in 24 well plates using the hanging drop method. Improved crystals of Howlett grew over ∼8 weeks in a 2 μl drop, which contained a 1:1 volume ratio of protein solution to mother liquor. The mother liquor contained 100 mm BisTris (pH 5.5), 200 mm ammonium sulfate, and 20% (w/v) PEG 3350. X-ray data were collected from a single cryocooled crystal with 25% PEG 3350 at the Diamond Light Source (Didcot, Oxon, United Kingdom) on an ADSC Q315 CCD detector on station I03 (λ = 0.97A). Diffraction images were collected at an oscillation angle of 1.0o. Data were processed in the monoclinic space group P21 using the HKL2000 package. Initial phases were obtained through molecular replacement using the program PHASER. The search models used for molecular replacement were the 1.68A structure of bovine chymotrypsin (Protein Data Bank code 4CHA) and a model of ecotin generated based on its sequence alignment with E. coli ecotin (code 1ECZ). model building was performed with Coot, and refinement was performed with REFMAC5. A set of reflections was set aside for Rfree calculation.
Enzyme activity assay3 Unit of enzyme activity (U) was defined as: 1mg of protein will hydrolyze 1.0μmol of N-benzoyl-L-tyrosine ethyl ester (BTEE) (ICN Biochemicals, Thame, UK) per minute at pH 7.8 at 25°C. Enzyme activity was determined by UV-Vis spectrophotometer (Shimadzu, Model 1601; Tokyo, Japan).
BTEE + H2O N-benzoyl-L-tyrosine + ethanol
The assay mixture was composed of 1.42ml of Tris-HCl buffer (80mM, pH 7.8), 1.4ml of 1.18 mM BTEE and 0.08 ml of 2M CaCl2. After addition of 0.1 ml of protein solution (all 0.1ml samples contained in the range of 195-205ng of protein as quantified by NanoDrop 2000), the reaction was carried out at 25°C for 3 minutes, measuring the absorbance of the solution at 30 second intervals at 256nm. ΔA was plotted against time, and the specific activity was calculated as follows from a linear segment of the curve:
![]()
where ΔA is the absorbance change of the solution at 256nm, P represents the amount of protein contained in 0.1ml of protein solution, and 0.964 is the molar extinction coefficient of N-benzoyl-L-tyrosine at 256nm. The same assay was carried out with 3-[({[({1-[(4-nitrophenyl)carbamoyl]-2-phenylethyl}carbamoyl)methyl] carbamoyl}methyl)carbamoyl]propanoic acid (ICN Biochemicals, Thame, UK), a larger substrate analogue, henceforth referred to as LCSA (larger chromogenic substrate analogue).
RESULTS
Wolverine possesses a protein with homology to the axolotl Amblox protein.
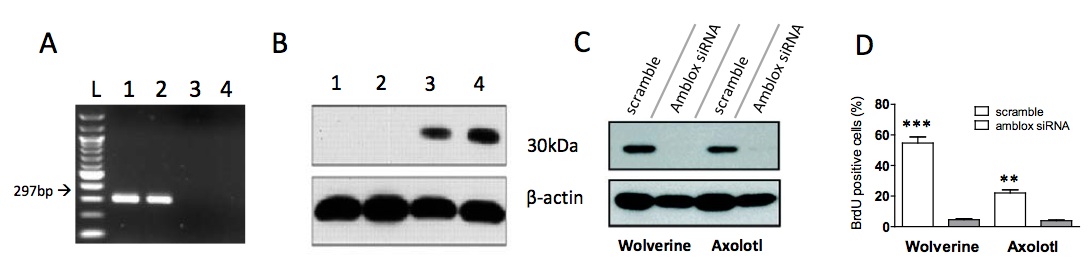
To determine whether Wolverine possesses a similar gene to the axolotl amblox, we first performed a standard PCR using primers specific for the amblox gene sequence downstream of the promoter. For PCR and protein assays, we harvested epithelial samples from both the axolotl and Wolverine, as well as normal human and other mutant controls (X-Men mutants possessing abilities other than Healing Factor). We observed a band for the 297bp PCR product in the axolotl and Wolverine samples (Fig. 1a). This band was absent in the samples from a normal human and from mutants possessing other superpowers, suggesting this gene is specific to Wolverine’s “mutation” and may be responsible for his Healing Factor.
Figure 1. [Click Image to Enlarge] Wolverine possesses a protein that is nearly identical to the axolotl Amblox protein. (A) 1.5% agarose gel electrophoresis of PCR products generated with primers specific for amblox gene. DNA was obtained from epithelial samples from an axolotl (lane 1), Wolverine (lane 2), a normal human (lane 3) and another mutant (lane 4). The first lane (Lane L) contains a 100bp ladder (NEB), lanes 1 and 2 showed the presence of a 297bp fragment of the amblox gene and lanes 3 and 4 are negative for the amblox gene. (B) Proteins were detected in cell lysates from epithelial samples using an α-Amblox antibody. The α-Amblox antibody can detect a protein in Wolverine (lane 4) and Amblox (lane 3). There was no protein detected in normal human and other mutant control (lanes 1 and 2). Beta actin was used as a loading control. (C) Effects of stable siRNA knockdown of Howlett and Amblox proteins. Epithelial cells from Wolverine and axolotl were cultured in vitro. Wolverine and axolotl cells were treated with amblox siRNA for 5h, followed by detection of protein content in cell lysates. (D) Cells transfected with scramble or amblox siRNA were incubated with BrdU for 2h post-transfection. BrdU positive cells were quantified by flow cytometry. There is a significant decrease in BrdU positive cells after siRNA knockdown (***p<0.001, **p<0.005).
Next we sought to identify whether an Amblox-like protein was present in Wolverine. We performed a Western Blot on cell lysates using a monoclonal antibody against the Amblox protein, graciously donated to us by the laboratory of Dr. Hank McCoy. We detected a protein in Wolverine cell lysates, which we have dubbed Howlett, at ~30kDa, of comparable size to the Amblox protein (Fig. 1b).
To confirm that expression of the Howlett protein is indeed a result of the gene found in Fig. 1a, we harvested skin cells from Wolverine and the axolotl and cultured them in vitro, which proved to be quite easy due to their regenerative capacity. We then transfected these cells with an siRNA construct against the amblox gene. Expression of both Amblox and Howlett decreased significantly upon knockdown with the amblox siRNA compared to scramble siRNA (Fig. 1c), confirming that the gene found in Fig. 1a, named howlett, was responsible for the Howlett protein expression. Proliferation of cells treated with amblox siRNA also exhibited decreased proliferation compared to those treated with control siRNA, as assessed by BrdU incorporation and flow cytometry (Fig. 1d).
Howlett is found in all tissues in Wolverine and at higher concentrations than Amblox is found in axolotl.
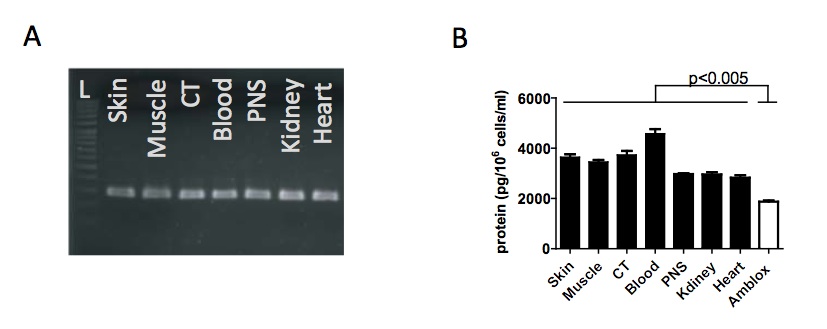
Wolverine appears to have the ability to heal any part of his body. To verify whether this is indeed the case, we biopsied numerous tissues from Wolverine and performed a PCR for the howlett gene, using the same primers as for Fig. 1a and a Western Blot using the same antibody as Fig. 1b. We found the howlett gene in all biopsied tissues (Fig. 2a). The Howlett protein was also found in all biopsied tissues at varying concentrations as assessed by ELISA (Fig. 2b). We also found that Howlett is expressed at significantly higher concentrations than Amblox (p<0005), which corroborates with the higher band intensity found in the Western Blot (Fig. 1b).
Figure 2. [Click Image to Enlarge] Howlett is found at higher levels in all tissues in Wolverine relative to Amblox in axolotl. (A) 1.5% gel electrophoresis showing PCR results for amblox/howlett gene in different tissue samples from Wolverine. The 297bp fragment was found in all tissues tested from Wolverine. L is a 100bp ladder. (B) Howlett protein concentration as measured by ELISA. Howlett was found to be expressed in all tissues in Wolverine at significantly higher concentrations than Amblox was expressed in axolotl. Protein concentrations were measured in 106 cells.
Howlett has increased enzymatic activity compared to Amblox protein
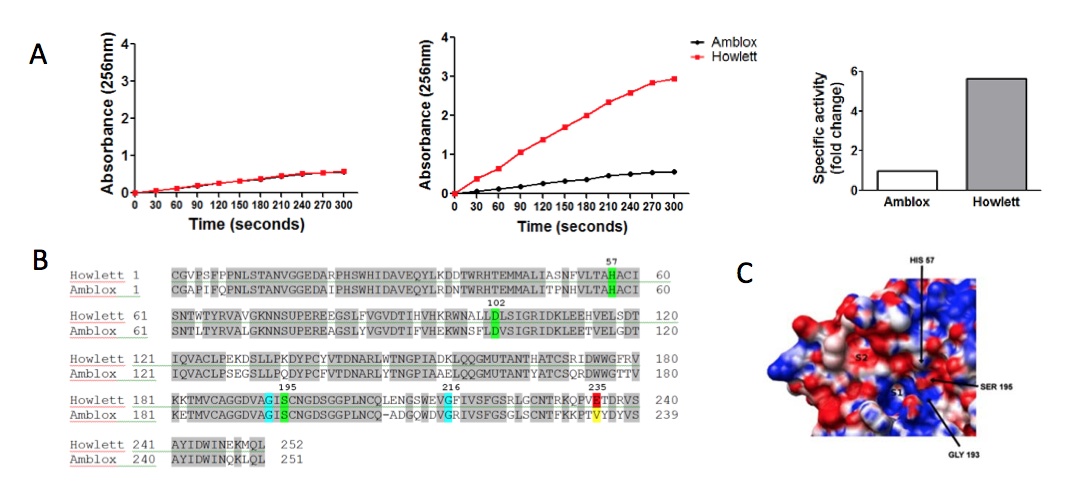
Although the axolotl is able to regenerate its limbs in 30 days and has improved wound healing relative to other animals, Wolverine has been shown to heal his wounds in a matter of minutes. To assess the reason for this accelerated healing process, we sought to determine the activity of Howlett within Wolverine’s cells. Wolverine has a significantly elevated amount of Howlett per cell than the axolotl by a factor of 100 (Fig 3a). However, this does not wholly account for the rate at which he can regenerate. To further assess this, we set up an activity assay using a panel of chromogenic substrates for various proteases. Howlett and Amblox cleaved BTEE (smaller substrate analogue) at comparable rates with comparable specific activities. With the LCSA (larger substrate analogue), we observed a higher rate of product formation by Howlett, with a 5.66-fold increase in specific activity over Amblox.
Figure 3. [Click Image to Enlarge] Howlett has increased specific activity attributed to a unique S2 “pocket” adjacent to the active site. (A) Purified enzyme was incubated with chromogenic substrates: (left) BTEE (small substrate) or (middle) LCSA (large substrate). Absorbance of cleaved substrate was measured at 256nm. The specific activity of each protein was calculated (see Materials and Methods for calculation) and is expressed as fold change over specific activity of Amblox (right). (B) Mass spectrometry results show full amino acid sequences of Howlett and Amblox proteins. There is 80% homology between Howlett and Amblox proteins. The catalytic triad (in green) is identical in the two proteins, as are the S1 pocket (G216) and the oxyanion hole (G193) (both in blue). At position 235, Howlett contains a glutamate residue (red) whereas Amblox contains a valine residue (yellow). (C) Close-up of the active site of Howlett. Substrate binding sites of Howlett: S1 and S2 pockets as well as the catalytic pocket as denoted by the catalytic triad.
Howlett has a pocket that can bind amino acids to efficiently cleave substrates
To shed light on the cause for increased enzymatic activity, we sought to determine the structure of Howlett and to compare it to that of Amblox. With the help of our collaborators at Lehnsherr University, we elucidated the sequences of Howlett and Amblox by mass spectrometry (Fig. 3b) and generated a 3-D structure from X-ray crystallography data (Fig. 3c). We observed that there is 80% homology in protein sequence between Amblox and Howlett. Notably, residues of the catalytic triad (in green) – H57, D102 and S195 – are identical . We also saw conservation of glycine residues at positions 193 and 216, which are integral parts of the oxyanion hole and S1 pocket, respectively. 3-D structure analysis revealed an S2 pocket comprising a non-homologous area that includes a glutamate residue at position 225, which is not present in Amblox.
DISCUSSION
In this study, we sought to determine the mechanism of Wolverine’s incredible regeneration capacity. Noting that Wolverine’s healing ability was reminiscent of limb regeneration seen in axolotls, we identified that Wolverine expresses a protein nearly identical to Amblox, the protein known to contribute to axolotl limb regeneration. We have dubbed this novel protein ‘Howlett’. We found that a monoclonal antibody specific for a region distal to the active site of Amblox was also reactive towards Howlett in Wolverine cell lysates from all tissues (Fig. 1e). This protein was found in greater concentrations than Amblox per 106 cells (Fig. 2a). siRNA knockdown assays confirmed that this protein contributed to increased replication and thus regeneration (Fig. 1c,d).
Although Howlett was found at higher concentrations than Amblox in comparable cell lysates, this difference did not entirely account for the increased speed at which Wolverine can regenerate compared to the axolotl. Hence, we sought to characterize the two proteins in order to determine the mechanism(s) by which the Howlett protein may be responsible for the increased regeneration capacity observed in Wolverine. To this end, we resolved the crystal structure and amino acid sequence of Howlett in order to elucidate any differences between the two proteins. The proteins were found to have nearly identical structures (80% homology), corroborating the binding of the α- Amblox antibody to Howlett. It has been previously described that Amblox is a serine protease with specificity similar to chymotrypsin, as it cleaves on the carboxyl side of large hydrophobic residues such as tryptophan, phenylalanine, and tyrosine . We confirmed that the Howlett active site is comparable to well-characterized serine proteases, such as 4trypsin, chymotrypsin and elastase, utilizing the mechanism of the catalytic triad4. With the high percent homology observed between Amblox and Howlett, we postulated that Howlett, like Amblox, could cleave chymotrypsin substrate analogues. Therefore, we assessed the specific activities of Amblox and Howlett with a selection of chromogenic chymotrypsin substrate analogues. Notably, the specific activities of Amblox and Howlett were comparable with the substrate analogue BTEE, a smaller chromogenic substrate. However, with the larger substrate analogue (LCSA), Howlett displayed a 5.66-fold increased specific activity over Amblox. Looking closely at the active site of the two enzymes and the surrounding regions, we noted some structural features that we hypothesize contribute to the results observed. Firstly, both enzymes contain an S1 pocket adjacent to the active site, like that seen in chymotrypsin. As with chymotrypsin and Amblox, this pocket serves as a binding site for the large hydrophobic residues at position 1(P1), adjacent to the peptide bond to be cleaved (scissile bond)4. Secondly, we noted a second pocket (S2) adjacent to the S1 pocket that includes a glutamate residue in the Howlett protein, but not in Amblox. This S2 pocket could be a contributing factor for the increased activity seen with Howlett and not with Amblox. We postulate that this glutamate functions to create an electrostatic interaction with a lysine residue on the amino terminal side of the large hydrophobic residue that binds at P1. The S2 pocket likely provides specificity for the substrate, and offers additional steric stabilization for positioning of the scissile bond into the active site. The increased activity of Howlett likely contributes a great deal to the heightened rate of replication in Wolverine compared to that seen in the axolotl. Further investigation is warranted to determine other mechanisms contributing to the observed increase in protein levels. These should include transcript and copy number analyses.
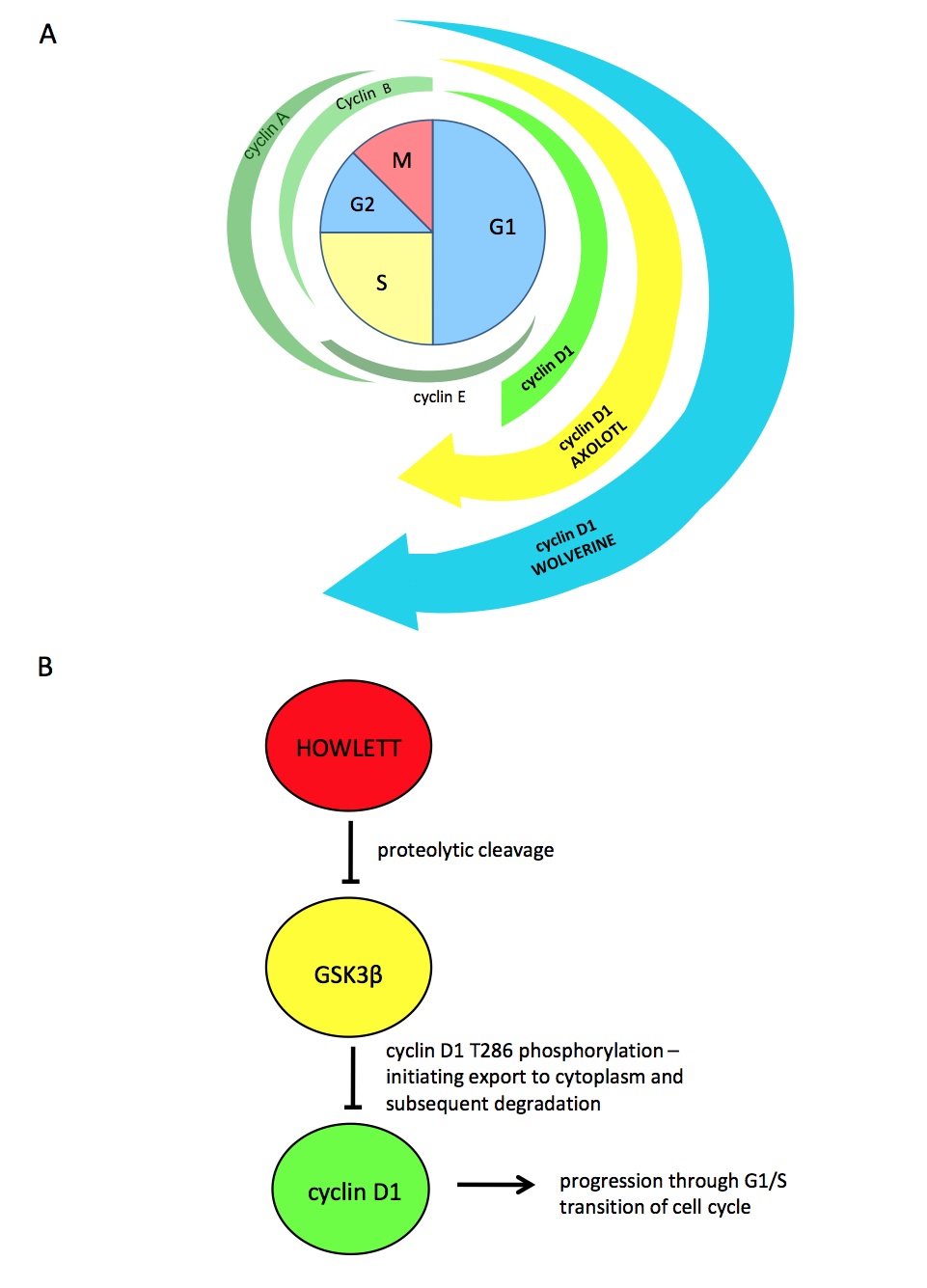
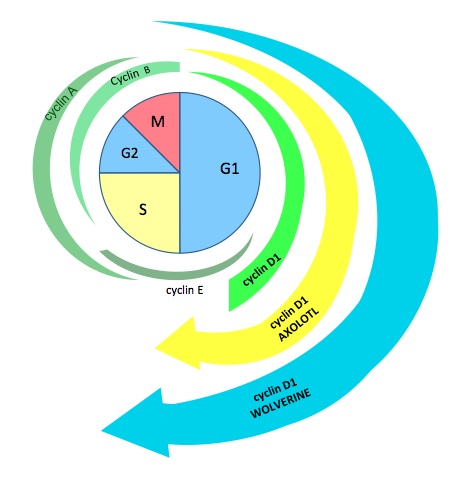
It is known that Amblox targets glycogen synthase kinase-3β (GSK3β)5 and that GSK3β has a function in cyclin D1 degradation6. Within the nucleus, cyclin D1 levels rise and interact with cyclin dependent kinase 4 (CDK4) or CDK6, which pushes the cell through the G1/S checkpoint. Cyclin D1 levels then fall and must rise again for another replication cycle to occur. GSK3β contributes to cyclin D1 degradation by phosphorylating cyclin D1 on T286, which facilitates its nuclear export by the exportin CRM1 to the cytoplasm where it is subsequently degraded6. Knowing this, we propose the following mechanism for the contribution of Howlett to Wolverine’s heightened regeneration capacity (Fig. 4). Howlett cleaves GSK3β, thus inhibiting GSK3β-mediated cyclin D1 degradation. This results in constitutively increased levels of cyclin D1, which continually push the cell through the G1/S checkpoint, thus initiating replication at a high rate. Preliminary data support this mechanism, as cyclin D1 levels analyzed at different stages of induced cell cycle arrest showed high levels at all stages of the cell cycle (data not shown).
Figure 4. [Click Image to Enlarge] Proposed mechanism of regulation of the cell cycle by the Howlett protein. (A) Cyclin levels in regular human cells are shown in green. We propose that cyclin D1 levels are increased in axolotls and further increased in Wolverine, accounting for the increased rate of replication and regeneration observed. (B) A proposed integration of Howlett into normal cell cycle regulatory pathways, accounting for increased levels of cyclin D1 with subsequent progression through the G1/S cell cycle checkpoint. We propose that Howlett inhibits the inhibitor (GSK3β-mediated degradation) of cyclin D1.
Our results have provided novel insight into the increased regeneration capacity observed in Wolverine. Further details of this pathway remain to be elucidated, particularly mechanisms that regulate the initiation and termination of regeneration at the appropriate times. To this end, the following questions remain. How is Howlett activated upon injury? Does it respond to a specific stress signal within cells at the injury site? Once regeneration is complete, how is Howlett inactivated? Is Amblox controlled by the same regulatory mechanism in the axolotl? Are both proteins inhibited/degraded? In vivo models will provide crucial insight into these mechanisms. Furthermore, we expect in vivo models will allow us to investigate the clinical potential of this highly active protein. For example, will Howlett impart regenerative capacity to injured tissues when injected into model organisms, or will it be immunogenic and initiate a detrimental immune response? Finally, are ‘template’ cells needed to be present in order for proper regeneration to occur? This question is particularly interesting in the context of transplantation where whole organs are removed.
The answers to these burning questions are highly sought-after because the regeneration capacity that the Howlett protein imparts in Wolverine could have substantial clinical implications. A vast array of human pathologies could benefit from a regeneration treatment modality. Howlett could potentially improve outcomes for patients suffering from spinal cord injury, severe burns, amputations, stroke, heart attack, and organ failure, to name a few. Given the incredible contributions the Howlett protein could have in the field of Regenerative Medicine, further investigation of this protein, its signaling pathway and regulatory mechanisms is certainly warranted.
ACKNOWLEDGEMENTS
We gratefully acknowledge all of our collaborators for their generous donations and insight. A big thanks to Aaron Huffsmith for being nerdy, we appreciate you bud.
COMPETING FINANCIAL INTERESTS
The authors disclose no competing financial interests.
REFERENCES
1. Shimokawa et al. 2012. Okajimas Folia Anat Jpn. 89(3): 75-81. 2. Kristensen et al. 2012. Nat Methods. 9(9): 907-909.
3. Hong et al. 2007. J Enzym Cat B. 45(2): 84-90.
4. Menten et al. 1987. Nature. 13(4): 56-72.
5. Menger et al. 2010. Fab World of Axolotls. 3(5): 34-52. 6. Alao et al. 2007. Mol Cancer. 6(24): 28-35.
FIGURE ACKNOWLEDGEMENTS
Figure 1a: Secka et al. 2011. Gut Pathog. 3(1):5.
Figure 1b: Deisenroth et al. 2010. Mol Cell Biol. 30(16): 3981-3983.
Figure 1c: Hou et al. 2011. Endocr Relat Cancer. 18(6): 687-697.
Figure 1e: Deguchi et al. 2009. PLoS One. 4(5):e5598.
Figure 2d: Fernandez et al. 2008. J Mol Biol. 377(5): 1488-1497.
In case it wasn’t clear already, this article is not real.