PRELIMINARY DATA FROM THE UNICORN GENOME: THE FIRST POSSIBLE INDICATION IN HISTORY OF MAMMALIAN EVOLUTION OF HYBRIDIZATION ACROSS ORDERS
Journal of Fantastic Phylogenetics 2012 8(9): 42-49
Authors: Jean-Pierre Marielle III & Justin Case | pdf version
In this study we present the preliminary finding of what could be the first record in history of hybridization between mammalian Orders (Perissodactyla and Cetacea). Phylogenetic analyses from the unicorn (Equus unicornus) mitochondrial genes (protein-coding Cytochrome b, Cytb) show a matriline belonging to the Narwhal (Monodon monceros) while nuclear (V-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog, Kit) clearly show a sister relationship of the unicorn-pegasus (Equus alatus) clade with the rest of the Perissodactyl family Equidae. Our preliminary study strongly suggests the need for further investigation because this finding could completely revolutionise our understanding of the processes of hybridization in mammals.
Introduction
The unicorn (Equus unicornus J.P. Marielle I) is a charismatic and iconic species of the Equidae (Peryssodactyla, Mammalia), forming with its sister species pegasus (Equus alatus S. Barrett) a clade sister to the wild horse and the donkey (Yule et al. 2001). This mysterious Equid is notoriously hard to study, and has been recorded sporadically appearing on all continents. The only known stable populations are recorded in the Arctic circle, Antarctica and the remote island of Tristan da Cunha (Klaus et al. 1993). Unicorns are one of the highest ranking endangered species alongside the Giant Panda (Ailuropoda melanoleuca David) and the Albino Alligator of the NYC sewers (Crocodilus newyorkensis H.G. Rogers), therefore recent studies have concentrated on its habitat, and how it might be affected by global climate warming (IUCN 2012). Future conservation strategies would greatly benefit from a deeper understanding of the evolutionary history of this species.
Today, as far as it is known, this species is threatened mostly by illegal poaching, historically the populations were decimated in Victorian times by the trend of collection of the natural world because of the alleged magical properties of its blood, and the ivory of the horn. This beast has been reduced to sparse populations, hardly spotted in the wild anymore and shrinking (Gold et al. 1991).
This species has been listed as a species of concern by the IUCN because of their low population numbers. The changing of global climate weather patterns is also considered a threat to unicorns because of their well studied and long documented association with rainbows, the occurrence of which will be compromised with shifting precipitation patterns (Ackbar et al. 2006).
The physiology of E. unicornus has always been an enigma mostly due to its chimeric appearance, as it is a mystery in its sister species E. alatus. Evolutionary theory always struggled with the apparent physical adaptations of this clade in the Equidae, so far that for some time the clade was called Chimerae (Mashruk 1972). In the middle of the 20th century there were some speculative studies suggesting the hybrid nature of these animals, however the evidence was never taken seriously and the data were weak. In the 1990s some study suggested the possibility of horizontal gene transfer through virus, however, these data were dismissed as far-fetched fantasies, recurring to a “near-religious illusion on systems science does not yet understand”, the harsh critique of the theory led to a fierce battle over the subject which continued for about a decade (Bond & Penny 2000).
Hybridization is widely known from the plant kingdom (Rieseberg & Carney 1998) and horizontal gene transfer has been repeatedly reported in prokaryotes (Jain et al. 1999). However, these forms of genetic exchange have never been prevalent (or even recorded in the case of horizontal gene transfer) in the Mammalia and never been recorded at such an evolutionary distance between organisms (Equidae and Cetacea).
Here are presented the incongruent results from two phylogenies built with mitochondrial and nuclear genes of E. unicornus showing relatedness to two different orders of the Mammalia.
Materials and Methods
Sampling
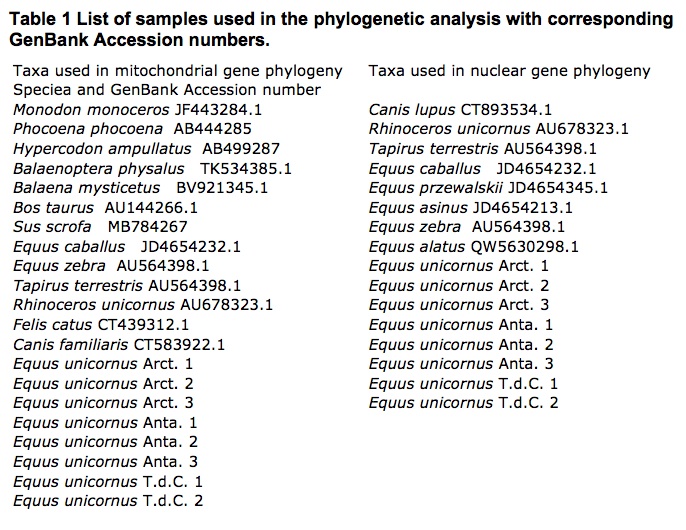
We examined a total of 7 E. unicornus from different populations: 3 from the Arctic, 3 from Antarctica and 2 samples from Tristan da Cunha. The rest of the sequence material used was downloaded from the GenBank website (samples are summarised in Table 1).
DNA extraction, sequencing, and mitochondrial and nuclear markers
We amplified sequences of two genes from exonic and intronic regions, one mitochondrial gene [Cytochrome b, Cytb (1140 bp)] and one nuclear gene [V-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog, Kit (273 bp; 1215 bp)] (Steiner & Ryder 2011). New sequences were submitted to GenBank (accession numbers in S1). PCRs were performed in a 20µL volume using Eppendorf Mastercycler Gradient thermal cyclers. Each reaction included 30ng of template DNA, 10µL of Taq buffer with 1.5 mM MgCl2 (Applied Biosystems), 0.3 µL of 10 mM deoxynucleoside triphosphates, 0.6 µL 1M of each primer, and 0.15 units AmpliTaq DNA polymerase (Applied Biosystems). PCR forward and reverse primers were designed in conserved exonic regions by using alignments of samples of Scrotifera mammalian sequences.
Sequences were assembled and edited using Sequencher 3.1.1 (Gene Codes, Ann Arbor, MI) software and then compared with GenBank using BLAST. Alignment was achieved using MAFFT (Katoh et al. 2002) and manual editing. Phylogenetic analyses were carried out using PAUP (Swofford 2003) and RaxML (Stamatakis 2006).
Phylogenetic analysis
Likelihood analysis: ModelTest 3.7 (Posada and Crandall 1998) selected a TIM + I + G model using Akaike weights, which we approximated in RAxML 7.0.4 (Stamatakis et al. 2005) with the GTRGAMMAI model of evolution. We ran the RAxML 7.0.4 likelihood analysis with 200 search replicates and 460 bootstrap replicates.
Results
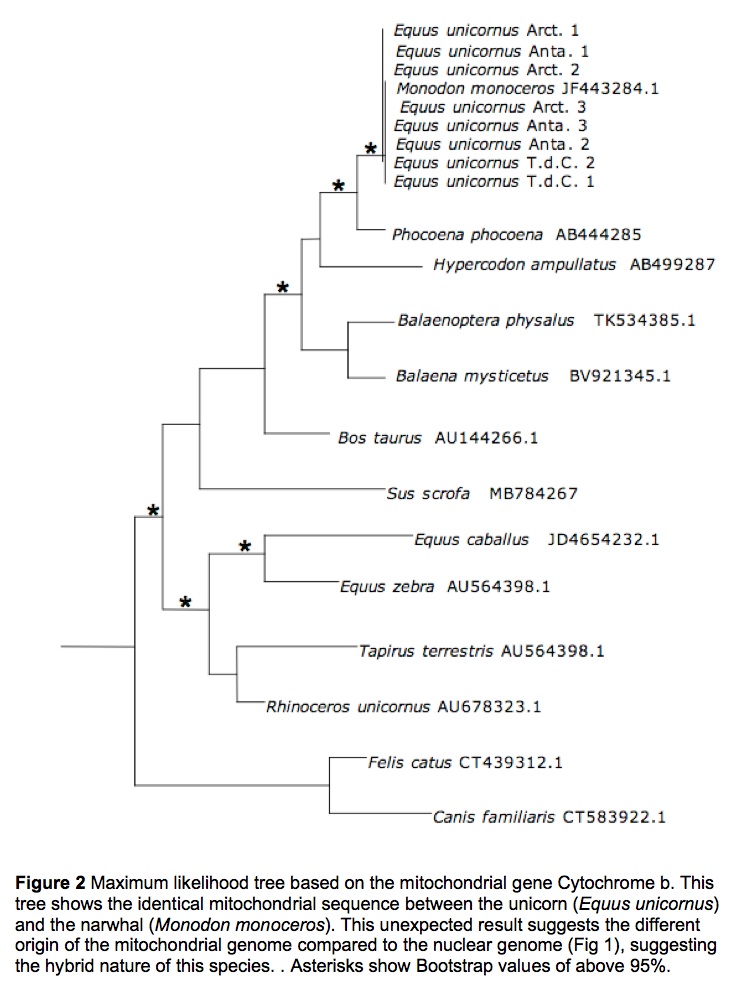
While the alignment of the nuclear genes did not show any problem, the alignment of the mitochondrial sequences proved to be difficult, if not impossible. A blast search of the sequences showed the samples of E. unicornus to belong to Monodon monoceros. The sample was thought to be contaminated, however unlikely, by narwhal DNA, and therefore the extraction was performed several times, producing each time the same sequence as result. This was beyond unexpected.
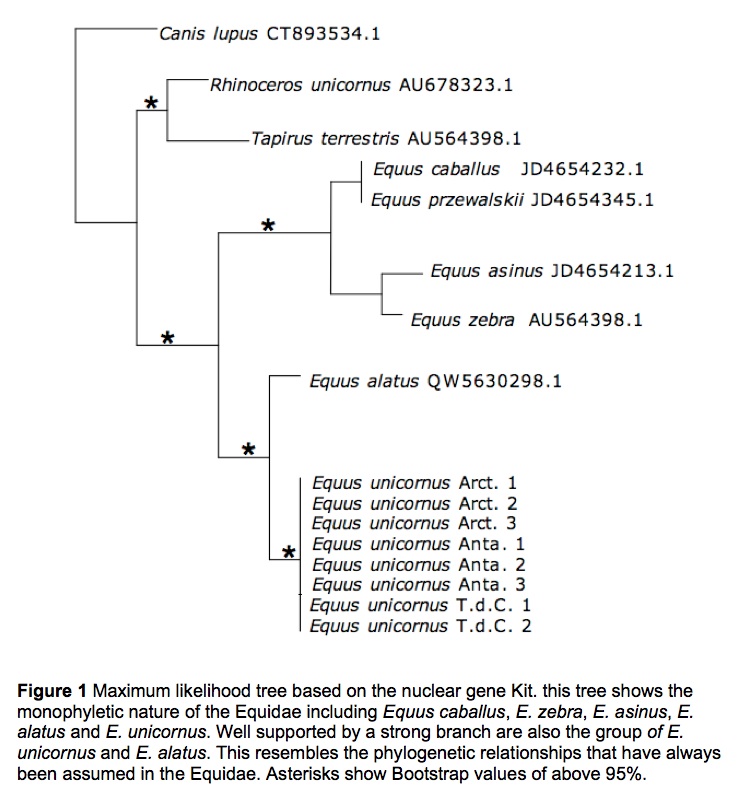
Two trees were produced with Maximum likelihood and sampling across the Scrotifera was carried out. The comparison between the alignments and phylogenetic analyses show a clear incoherence. Without the shadow of a doubt, the conflicting phylogenies between nuclear (Fig. 1) and mitochondrial (Fig. 2) genes suggest the acquisition of the mitochondrial gene possibly through the process of hybridization.
Discussion
Hybridisation
Hybridisation of closely related species has happened in several mammalian lineages; the black bear and the polar bear have shown hybridisaiton (Edwards et al. 2011; Miller et al. 2012), the Equidae themselves show several instances of hybridisation histories (Steiner & Ryder 2011). This is the first time that a hybridisation between species so far apart evolutionarily has been recorded. This finding needs to be confirmed by further research involving other genes and regions and including samples, that were unavailable for this study, for mitochondrial gene sequences from E. alatus.
Unfortunately the lack of a fossil record prevents us from investigating the evolutionary age of these hybridisation episodes any further than speculation. The application of a molecular clock could be a very rough estimate. The lack of fossil record and the limited access to samples and populations also limit the possibility of investigating if this is a single episode or hybridisation has occurred more than once.
Physiological considerations
When Marielle first described E. unicornus he was fascinated by the appearance of the horn and immediately made the connection with M. monoceratus. He was able to investigate the nature of the horn, from the carcass he was basing the his description on: the first record of a unicorn (aside from visual representations). When describing the attachment of the horn he stressed his uncertainty on where the horn was developing from, in fact, he could see a ridge of hardened bone along the nose. Now we know from observing the extant colonies that the horn is a tooth, which grows along the nose and at the height of the eyes changes orientation in development and at puberty will protrude from the animal’s forehead (Rudolf & Noel 2002). What seems to be puzzling at first is that only male narwhals will develop the horn, in the case of unicorns this will not occur; furthermore, considering the hypothesis of the matriline belonging to the narwhal, the sex chromosome Y would not be passed to the unicorn in this hybridisation event. One possible explanation would be that in the narwhal it is the chromosome X carrying the gene to code for the horn phenotype and it activated by the Y chromosome, while in the unicorn it is activated in both sexes.
It seems that if the E. alatus rose as a species in the same way as E. unicornus, then the nature of the wings would resemble that of the species that hybridised with the ancestor. In the 1970s Zev Mashruk, a prominent and active mammalian physiologist, published on the resemblance between the wings of the pegasus and the pelican (Mashruk 1973). This account was however dismissed due to his notorious and vastly self-advertised, extensive use of psychoactive drugs. Mashruk’s theory, so ridiculed, could be re-evaluated in the light of these new findings.
Conclusion
Further work is strongly suggested because of the highly significant implications of the finding: never it has been recorded such exchange of genetic material between such evolutionarily distant taxa.
Further research should include work on E. alatus as well as E. unicornus, addressing the question regarding the nature of this clade, specifically if this particular lineage is especially prone to this new type of hybridisation.
Acknowledgements
We would like to thank the Rainbow Foundation, Santa’s Workshop, Foxy Luddite Co-op for financial support. Thanks also to the Buddha, Old Ben Kenobi and our anonymous editor for helpful feedback on the manuscript. If you haven’t figured it out already, this paper is not real. Courtesy of Anna Bazzicalupo
Cited Literature
Ackbar, G., Solo, H., & Skywalker, L., 2006. The TRAP of Climate Change. Wookie Journal of Climate Patterns, 45: 23-28.
Anonymous, 2012. IUCN RedList available online at: http://www.iucn.org/about/work/programmes/species/our_work/the_iucn_red_list/. Accessed on October 27 2012.
Bond, J. & Penny, M., 2000. The Mystery of the Unicorn. Goldfinger Press Ldt., London.
Edwards, C.J., Suchard, M.A., Lemey, P., Welch, J.J., Barnes, I., Fulton, T.L., Barnett, R., O’Connell, T.C., et al., 2011. Ancient hybridization and an Irish Origin for the Modern Polar Bear Matriline. Current Biology, 21: 1251-1258.
Gold, F., Bond, J., & Penny, M., 1991. Unicorn-Human relations in history. South Pole University Press, Antartica.
Jain, R., Riviera, M.C. & Lake, J.A., 1999. Horizontal gene transfer among genomes: The complexity hypothesis. Proceeding of the National Academy of Science, 96:3801-3806.
Katoh K, Misawa K, Kuma K, Miyata T (2002) MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Research 30: 3059-3066
Klaus, S., Rudolf, R.N.R., Prance, E.R., & Noel, E.L.F., 1993. Chapter 11: Natural Populations of Unicorns. From: Sled-hunting rare animals: new perspectives on recording wild animal populations. South Pole University Press, Antartica. Pp. 568-598.
Marielle I, J.P., 1867. Animaux fantastiques. Vol. 2 p. 42
Mashruk, Z., 1972. A view from the clouds: perspectives on the Equidae and Chimerae. High Times in Science, 16:253-260
Mashruk, Z., 1973. Flying between the Pegasus and the Pelican. Acid Musings, 42:54-76.
Miller, W., Schster, S.C., Welch, A.J., RAtan, A., Bedoya-Reina, O.C., Zhao, F., Kim, H.L., Burhans, R.C:, Drautz, D.I., Wittekindt, N.E.,Tomsho, L.P. et al. 2012. Polar and brown bear genomes reveal ancient admixtueand demographic footprints of past climate change. PNAS published ahead of print July 23, 2012, doi:10.1073/pnas.1210506109
Posada, D., and Crandall, K.A. 1998. MODELTEST: testing the model of DNA substitution. Bioinformatics, 14(9): 817–818.
Rudolf & Noel 2002 Physiology and development of the frontal tooth of the unicorn. Journal of Fantastic Physiology, 9: 235-242.
Rieseberg, L.H. & Carney, S.E., 1998. Plant Hybridization. New Phytologist, 140: 599-624.
Stamatakis A (2006) RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22: 2688-2690
Steiner, C.C. & Ryder, O.A., 2011. Molecular phylogeny and evolution of Perissodactyla. Zoological Journal of the Linnean Society, 163:1289-1303.
Swofford DL, 2003. PAUP*. Phylogenetic Analysis Using Parsimony *(and other Methods). 4 ed. Sinauer Associates, Sunderland, Massachusetts.
Yule, C., Rudolf, R.N.R., Klaus, M.R.S., Prance, E.R., Noel, E.L.F., & Klaus, S., 2001. An unclear nuclear gene phylogeny of the Equidae. Journal of Fantastic Phylogenetics, 34(7): 56-60.