THE HUMAN GENOME PROJECT: THE IMPACT OF GENOME SEQUENCING TECHNOLOGY ON HUMAN HEALTH
(August 2003)
What is the Human Genome Project?
Genome sequencing technology has led to many recent scientific breakthroughs. These breakthroughs have captured the interest of the public and are being reported with excitement by both the media and scientific journals. The completion of the human genome project (HGP) is an example of newsworthy science that has the potential to have major effects on our society today. The HGP was an initiative started in the early 1990’s that has involved the efforts of hundreds of scientists to generate high-quality reference sequence for the 3 billion base pairs of nucleotide sequence that make up the human genome. The complete string of nucleotide letters that make up the DNA sequence in our cells is often referred to as our genome. This DNA sequence contained in a genome contains the complete code that determines which genes and proteins will be present in human cells. By reading the sequence of the human genome, scientists hope to gain an understanding of the underlying code that determines how a complex biological system, such as a human cell, acts and reacts. Insights from deciphering the human genome have potential to be applied to a better understanding of human health and could help to develop better treatments for disease.
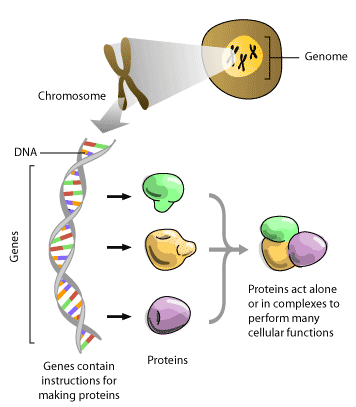
The HGP started at a meeting of scientists during which the value of knowing the genome sequence of an organism was recognized. Officially, funding for the project began in the 1990 with the goal of sequencing the human genome by 2005. At that time, many scientists thought this goal was out of reach. Nonetheless, scientists at genome centres took up the challenge. From the beginning, the project has also played a large part in driving the development of technology that aided the high-throughput sequencing of genomes from other model organisms such as mouse, worm and yeast. By comparing sequences from these different model organisms, scientists gain a better understanding of the important pieces of code in genomic DNA sequence since conservation of sequences between two organisms that diverged phylogenetically millions of years ago, like humans and worms, implies that the conserved sequence is important for function. The HGP has also played a role in developing the computational resources and expert personnel necessary for handling data generated on a genomic scale. Ten years after the official beginning of the HGP, the first working draft of the human genome was announced. Two large groups of scientists published the first analyses of this human genome sequence in the February 2001 issues of the journals Nature [1] and Science [2]. The race to publish human genome sequence information was fuelled by competition between research from the publicly funded HGP and the privately owned company, Celera Genomics. The public effort produced results which were freely available whereas the data from Celera was available for a fee. This first working draft of the human genome sequence was hailed with much excitement and fanfare as the “completion of the human genome” in the media. However, this first draft was not considered to be complete by scientists because of significant gaps in the sequences (this draft was 90% complete). For scientists, the high-quality reference sequence publicly released in April 2003 represents the first real step to having “finished” human sequence on hand (this draft represents sequence information that is considered to be 99% complete) [3].
What have we learned from the Human Genome Project?
These major accomplishments in genome sequencing provide a wealth of information that aid in the understanding of basic biological processes. With genome sequence in-hand scientists are now more effectively able to study gene function and explore new areas of research such as how human variation contributes to different diseases worldwide. Scientists today are discovering that the more we learn about the human genome, the more that there is to explore. For instance, as a first step in understanding the genomic code we have learnt that the human genome is made of 3.2 billion nucleotide bases (of which there are four types: A, C, T, G). It is thought that over 30,000 genes are encoded by this sequence. Yet we have also discovered that over 50% of the human genome is repetitive sequence that does not code for any proteins and the function of this large portion of “junk” DNA is still puzzling scientists. Along similar lines, the HGP has shown us that the average length of an expressed gene is 3000 bases long. Genome sequence information has helped scientists more easily identify candidate disease genes, however, we also realize that over 50% of the genes discovered in the human genome are still classified as having unknown function. Human genome sequence information reveals that genome sequences from person to person are almost (99.9%) identical. Interestingly, comparative genomics shows 95% sequence similarity between the human and chimpanzee genomes. Scientists are just beginning to understand how this small amount of variation contributes to differences in disease incidences in different populations. The discovery of about 3 million locations that have single base differences in the human genome (called single nucleotide polymorphisms or SNPs) offers insights into how genomic information could be used to discover information related to the incidence of common human traits, including susceptibility to certain diseases and illnesses.
The HGP has also shown us that the powerful methods of genome sequencing technology raise important ethical and policy issues for individuals and society. Access to genome sequence information, privacy related issues and the appropriate use of this sort of information are all important issues for researchers, governments, and policy makers worldwide. The HGP has great potential to benefit society. An understanding of human variation could be directly translated to human health with the creation of better treatments and personalized medicine. In our complex world, it is also important that human genome information be protected. Scientists, policy makers, educators and ethicists have recognized the need to encourage dialogue with the public about human genome sequence information and potential implications. A number of excellent resources are available online which celebrate human genome based discoveries and provide information about the implications of genomics in today’s society [4].
How is genome sequencing technology helping aid research today?
An example of how genome sequence technology can make a difference in today’s society is illustrated by the recent outbreak of severe acute respiratory syndrome (SARS) worldwide. Several hundred cases of severe atypical pneumonia were reported in Guangdong Province, China in late 2002. In March 2003, SARS had spread to healthcare workers in Hong Kong and a worldwide epidemic was in the works. By late June, 250 Canadian cases of SARS had been reported to the World Health Organization (WHO) and 38 patients had died as of July 10th, 2003 [5]. New clusters of patients, including healthcare workers, in Toronto drew a lot of attention from the media worldwide. An isolate of a coronavirus obtained from the second patient in Toronto, called Tor2, was identified as the SARS virus. The virus was then sent to the British Columbia Centre for Disease Control in Vancouver for genome sequencing by the Genome Sciences Centre at the BC Cancer Agency [6]. The virus sample arrived on April 2nd and by April 11th the genome of this virus had been sequenced. The first complete assembly of the SARS viral genome was deposited into public sequence databanks the next day on April 12th. The information derived from the genome sequence of SARS gave insights into the origin of the disease as well as aided with patient diagnosis. The speed at which this 29,751 base genome was successfully sequenced is an exciting accomplishment for the field of bioinformatics. Fortunately, the SARS epidemic worldwide is now under control and scientists now have a better understanding of the disease, due in part to the knowledge derived from genome sequencing. It was less than one month, from the isolation of the virus to the publication of the sequence of the SARS genome [7]. This amazing pace of current research can be attributed directly to the development of the technology and expertise emerging out of the HGP and serves to illustrate how genome sequencing technology and bioinformatics will benefit our basic understanding of life and disease processes.
References
1. Nature. 2001 Feb 15; Issue 409(6822)
2. Science 2001 Feb 16; Issue 291(5507)
3. NCBI Map Viewer
4. Genomics and Its Impact on Science and Society
5. World Health Organization
6. Genome Sciences Centre SARS page
7. Science. 2003 May 30; 300(5624):1399-404.
(Art by Jiang Long – note that high res versions of image files available here)