WOOKIEE FOREPLAY: TRANSGENERATIONAL EPIGENETIC REGULATION OF FORCE-SENSITIVITY BY ELABORATE DUET SONG
Just another reminder that this paper will be published in print format in the fall, along with selected SCQ pieces from the last 7 or so years. If you want to submit your creative science pieces with a mind to make the first issue of this new journal, then please visit here for more information.
Annals of Praetachoral Mechanics. (2014). Vol 1. pp59-69 pdf download
ABSTRACT
Wookiees (W. wookiee) are monogamous, sentient mammals that have traditionally produced long and elaborate duet songs. It has been repeatedly proposed that duet mating songs for Wookiees might function to improve the fitness of offspring. In this study we aimed to compare DNA methylation of genes that may influence force-sensitivity in Wookiee pups from singing and non singing parents. Here we report that prenatal exposure to duet song altered the offspring epigenome. We identified 390 differentially methylated genes by genome-wide methylation profiling technology. We found five force-sensitivity genes (UTFL, ASFYL, LSGB, and NTRP) that were differentially methylated in pups from duet singing parents and confirmed that altered methylation of ASFYL and HMOWK affects gene expression. Furthermore, pups from duet singing parents had more midi-chlorians per cell, indicating a greater awareness for the Force. Taken together, our findings demonstrate that Wookiee mating song behaviour during prenatal development has a positive benefit on offspring.
Keywords: Wookiee, song, force sensitivity, epigenetics, DNA methylation
Introduction
A number of animals in the galaxy are known to produce elaborate mating songs, most notably species of monogamous birds and a few arboreal mammals, mainly Ewoks (E. ewok) and Wookiees (W. wookiee). The functions most frequently proposed for duet songs include territorial advertisement and strengthening pair bonds (Geissmann, 1999; Haimoff, 1981). Prenatal exposure to duet songs is believed to improve the fitness of the developing offspring, but this has not yet been demonstrated in any species. Previous efforts in studying the effect of duet songs have been conducted in Ewok and been largely unsuccessful. This is mainly attributed to fact that Ewok are extremely uncooperative and mischievous subjects. Wookiee on the other hand, are much better subjects due to their renowned intelligence and a propensity for force-sensitivity that is closer to human than most creatures. In fact, there have been several adept Wookiees of the Force, notably Lowbacca. Despite their fearsome appearance, Wookiees have a natural gift for musical song and are surprisingly gentle and sensitive creatures if not provoked. The Wookiee mating call is an old tradition of the Kashyyyk, but sadly knowledge of the culture has been fading over many generations.
Previously, we speculated that the advent of a superior specimen with heightened force-sensitivity is largely determined by epigenetic transmission. Perhaps the most prominent example of this phenomenon is the birth of Luke and Leia from Queen Padmé Naberrie Amidala. Many scholars attribute the differences in their upbringing as to why Luke realized the Force much earlier in life than his twin sister. Hallmark findings in the past have established a direct relationship between the number of midi-chlorians per cell and sensitivity to the Force (Russell and Rocheleau, 2014). Recent findings suggest that epigenetic modifications established in the prenatal environment can have a life-long effect on midi-chlorian content.
Given that Wookiee couples that produce duet songs are believed to produce healthier offspring, we hypothesized that prenatal exposure to duet singing may also enhance force sensitivity. To compare the differences between pups from singing and non singing parents, we conducted a genome-wide methylation analysis to determine differentially methylated genes associated with force-sensitivity. Here we provide compelling evidence that Wookiee pups conceived from parents that produce duet mating songs have a great number of midi-chlorians, and hence are more sensitive to the Force, than pups from parents that have forgotten the ritual.
Materials and Methods
Wookiee pup sample collection.
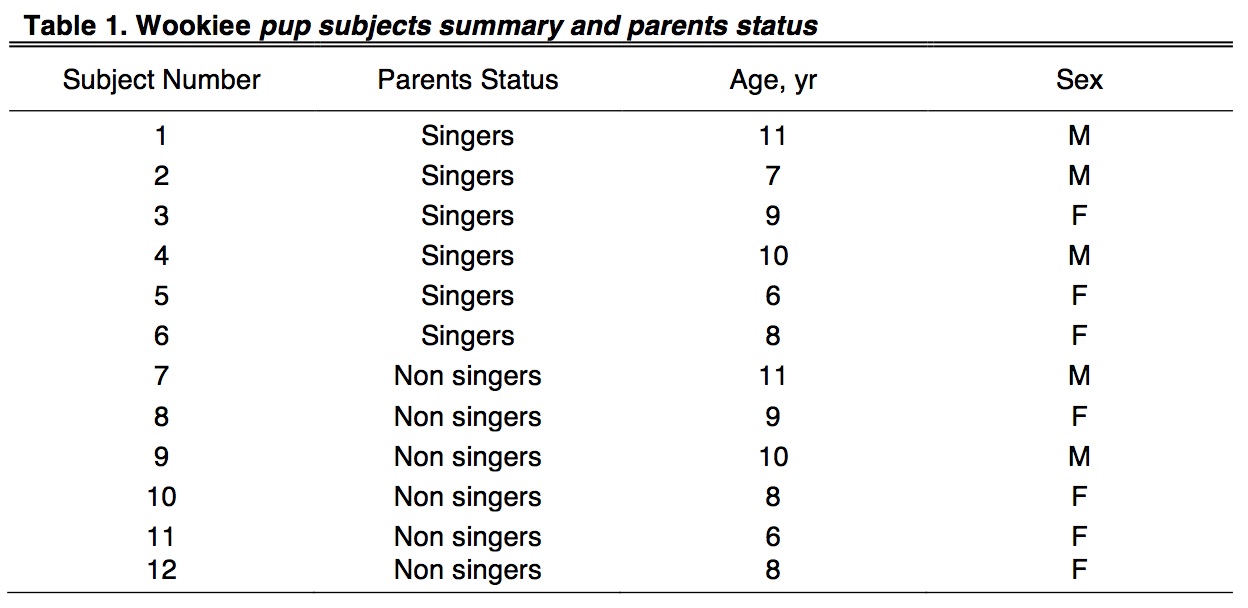
Blood samples were collected from healthy Wookiee pups at the Rwookrrorro nursery ring by standard phlebotomy techniques after receiving written consent from their parents. Subjects were selected between the ages of 6-11 years and were identified as generally healthy and without a diagnosis or history of any force-deficiency disorders. After venipuncture, EDTA was added as an anticoagulant and samples were stored at −80 °C until use.
DNA extraction and hybridization to Illumina Infinium assay.
Genomic DNA was extracted and purified from whole blood using DNeasy blood and tissue kit (Qiagen, Valencia, CA) according to manufacturer’s protocol. DNA was quantified by NanoDrop 2000 Spectrophotometer (NanoDropTechnologies, Inc.) before transfer to 4°C for short-term storage or -20°C for long-term storage. A bisulphite conversion reaction was performed using 500 ng of genomic DNA according to manufacturer’s instructions for the Zymo EZ DNA Methylation kit (Zymo Research, Irvine, CA). The bisulfite-converted sample was then hybridized to the WookieeMethylation27 BeadChip (Illumina, San Diego, CA) and DNA methylation was evaluated at the Arkanis’ Obi-Wan Genome Sciences Centre (Tatooine, Outer Rim Territories).
Analysis of differentially methylated genes.
Results from the Illumina Infinium array were interpreted with GenomeStudio software. The WookieeMethylation27 BeadChip quantifies methylation levels at 27,578 CpG loci spanning 14,495 genes (Rev. by Gupta et al. 2010). The methylation status of CpG sites are calculated as the ratio of fluorescent signal from one allele relative to the sum of both methylated and unmethylated alleles. The resulting average β-intensity signals range from 0 (unmethylated) to 1 (fully methylated) to represent the percentage of methylation. Differential methylation was defined as a significant difference in percentage of methylation between pups from duet singing vs. non singing parents, where a difference in percentage of methylation was ≥33.3% or ≤-33.3%, combined with an ANOVA P value of <0.05. ANOVA P values were calculated using GraphPad Prism 6 software (GraphPad Software, Inc. La Jolla, CA). To test the potential influence of age, an additional statistical analysis was performed with a mixed model ANCOVA to consider age as a covariate (Partek). Array data have been submitted to the Sentient Genetics Omnibus repository (SGO) and are available under accession number SGO126584.
List of differentially methylated force-sensitivity genes.
A list of genes encoding proteins within the force sensitivity pathway was curated using the JEDI pathway database (episode 6.0). The resulting list was compared against results from the Illumina methylation array to identified genes with altered methylation in pups from duet singing and non singing parents.
Real-time PCR.
Total RNA was isolated from whole blood using the Tempus Spin RNA Isolation Kit according to manufacturer’s instructions (Applied Biosystems, Foster City, CA), followed by quantification by NanoDrop 2000 Spectrophotometer (NanoDropTechnologies, Inc.). First-strand cDNA synthesis was carried out using the TaqMan Reverse Transcription Reagents (Applied Biosystems) with random hexamers according to the manufacturer’s protocol. Triplicate samples for quantitative real-time PCR were run in the Applied Biosystems 7500 real-time PCR System using the Power SYBR Green PCR Mastermix (Applied Biosystems). Each reaction contained 1 μl of cDNA in a total volume of 20 μl. Results were normalized to β–actin transcript levels.
Quantification of midi-chlorian DNA copy number.
Midi-chlorians were enriched from whole blood samples using the Midi-chlorian extraction kit (SithTech, Tatooine, Outer Rim Territories) according to manufacturer’s protocol. McDNA was extracted by digestion with Proteinase K (100 μg/ml) in a buffer containing 50 mM KCl, 10 mM Tris-HCl, 2.5mM MgCl2 and 0.5% Tween-20. Samples were incubated overnight at 37˚C then boiled for 5 min. McDNA was linearized by digestion with ChewIII enzyme. McDNA content was measured by a multiplex real-time PCR method as previously described (Andreu et al. 2009). Primers to amplify a single copy mcDNA gene R2D2 and a mcDNA TaqMan probe were used (Applied Biosystems).
R2D2 forward primer: 5’-CAGAATGATATTTGTCCTTCA-3’
R2D2 reverse primer: 5’-GATATGAAAAACCATCGTTG-3’
R2D2 probe: 5’-TGCCAGCCACCGCG-3’
McDNA quantity was corrected by simultaneous measurement of a single copy nuclear gene C3P0 gene using a commercial (Applied Biosystems). Real-time PCR reactions consisted of 1 μL of pre-developed assay regent for C3P0 and 112 nM of each mcDNA primers and mcDNA probe, and 2–10 ng of total genomic DNA extract. Thermocycling was completed out on an Applied Biosystems 7500 real-time PCR System with the following conditions: 2 min at 50 °C and 10 min at 95 °C, followed by 40 cycles of 15 s of denaturation at 95 °C and 60 s of annealing/extension at 60 °C. Calibration curves were generated to quantify mcDNA and nuclear DNA copy numbers, which were based on the linear relationship between the crossing point cycle values and the logarithm of the starting copy number (Phillips, 2014).
Results
Prenatal exposure to Wookiee duet song impacts the DNA methylation profile of hundreds of genes.
Six pups from singing parents and six from non singing parents were recruited from the Rwookrrorro nursery for this study (Table 1). Based on personal communication and interviews with the parents, we were able to formulate a composition of a typical mating duet (Fig. 1A), as well as trace sonograms to describe the distinctive calls produced during a song (Fig. 1B). Overall, the duet mating song is an elaborate custom with a well-defined sequence of howls, barks and intermittent screams.
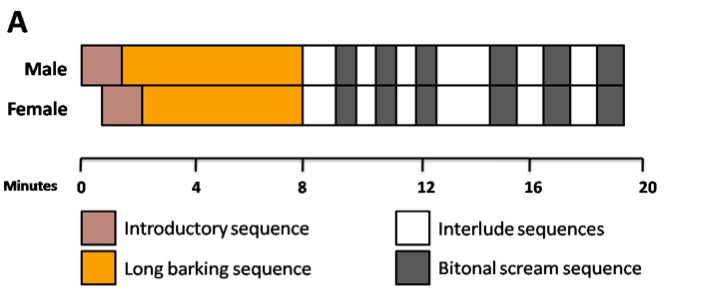
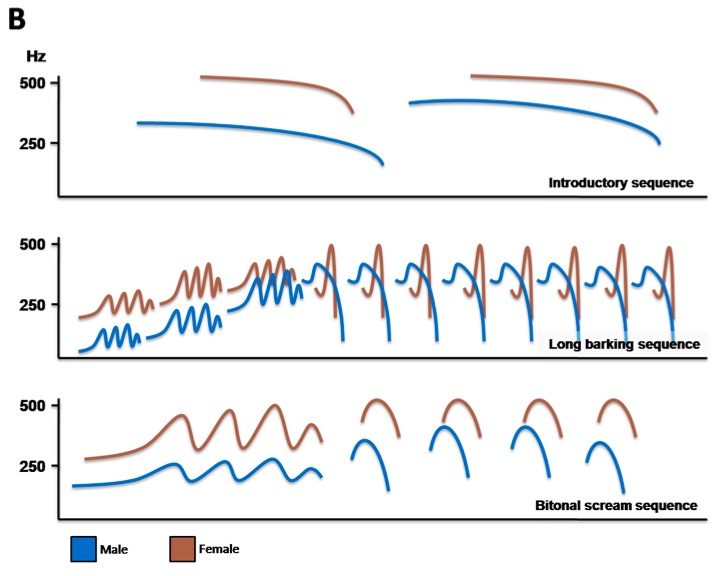
Figure 1 (Click to Enlarge). Duet mating song produced by adult Wookiee pairs. A: Schematic diagram illustrating the sequential nature of an entire mating duet. B: Sonagram tracings showing the types of call sequences produced during a song.
Using DNA isolated from whole blood samples, we used Illuminia Infinium technology to conduct genome-wide DNA methylation profiling. By a comparative statistical approach, we found that many genes are differentially methylated as a result of prenatal exposure to the duet song. We identified 390 genes showing differential methylation between pups from singing and non-singing parents (Fig. 2A). Of all the differentially methylated genes, 328 were hypomethylated and 62 were hypermethylated. In general, hypomethylation of gene promoters is associated with more robust transcriptional activity. Based on these findings, it appears that numerous genes are more active in pups from parents that sing duets.
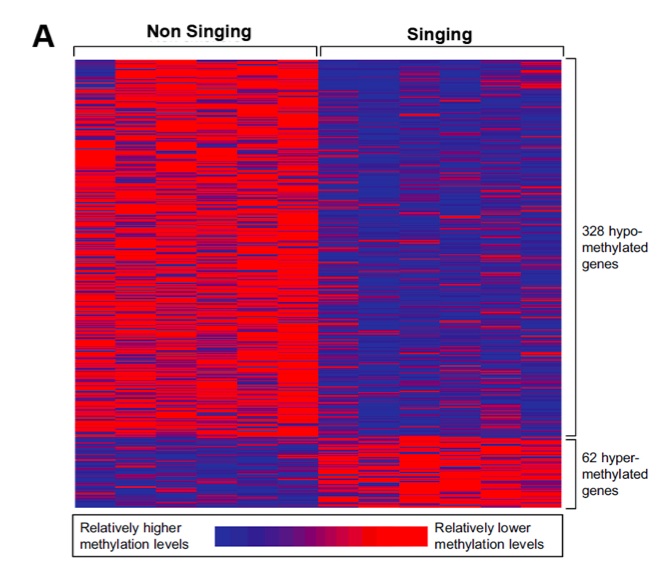
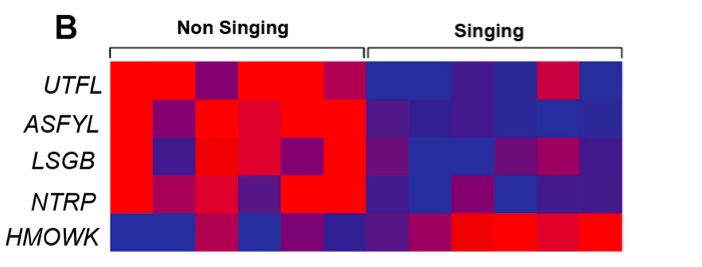
Figure 2 (Click to Enlarge). DNA methylation levels in pups from duet singing and non-singing parents. The relative methylation of genes are displayed with heat maps for the 390 genes differentially methylated in pups from singing parents (A). 5 genes detected as differentially methylated in our array, are force-sensitivity genes (B).
Force-sensitivity genes are differentially methylated in pups from duet singing parents.
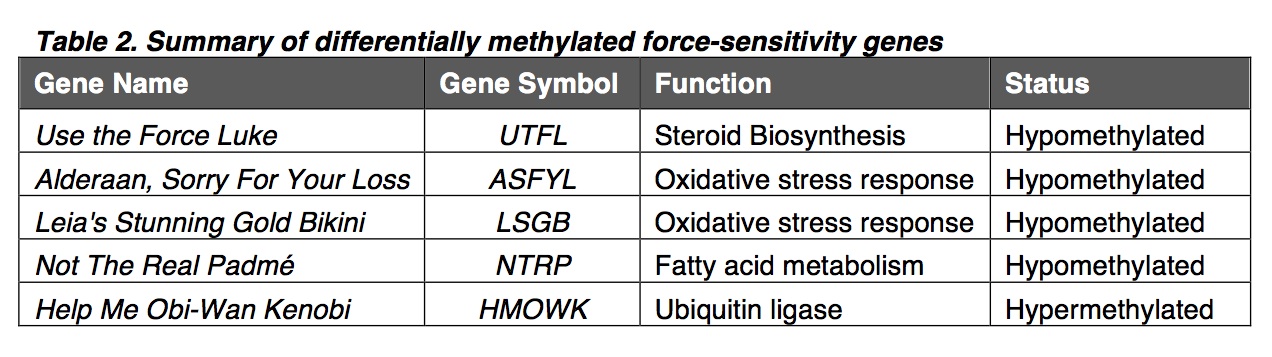
To focus on genes encoding proteins in the force-sensitivity pathway, we used the JEDI pathway database (episode 6.0) to generate a list of genes to be examined for differences in DNA methylation status. Comparing the list to our methylation array, we identified five genes that were differentially methylated in pups from parents that sing and do not sing (Fig. 2B). Of the differentially methylated force-sensitivity genes, four genes were hypomethylated (UTFL, ASFYL, LSGB, NTRP), and one was hypermethylated (HMOWK; Table. 2).
Methylation of ASFYL and HMOWK promoters affects gene expression.
An additional statistical analysis (ANCOVA) was performed for the selected force-sensitivity genes, where array-based methylation levels in singers were compared against non-singers while adjusting for age as a covariate. UTFL, ASFYL, and NTRP remained statistically significant (P < 0.05); however, the P values for LSGB and HMOWK became non significant and increased to 0.098 and 0.067 respectively. Nevertheless, the difference in methylation in HMOWK between singers and non-singers (Fig. 2B) is still biologically relevant. Among the differentially methylated force-sensitivity genes, two of them, mainly ASFYL and HMOWK are known to play a strong role in the control of force-sensitivity in humans. Previously, these genes have been implicated to be a determinant of longevity in Wookiees. ASFYL encodes a protein that regulates oxidative stress responses, and increased expression of ASFYL is associated with improved removal of reactive oxygen species (ROS) that are detrimental to cells. HMOWK encodes an ubiquitin ligase to the midi-chlorian outer membrane that has been proposed to play a role in the control of midi-chlorian morphology by regulating contacts with the endoplasmic reticulum and essential GTPases. This in turn promotes fusion of midi-chlorians as a mechanism to maintain midi-chlorian numbers in response to intrinsic signals.
We confirmed that DNA hypomethylation of ASFYL and hypermethylation of HMOWK affects gene expression by quantitative real-time PCR. As predicted, hypomethylation of ASFYL was associated with increased expression of ASFYL transcripts in pups from the singing parents group (Fig. 3A). Similarly, hypermethylation of HMOWK was associated with decreased HMOWK transcript levels in pups from the non singing group (Fig. 3B).
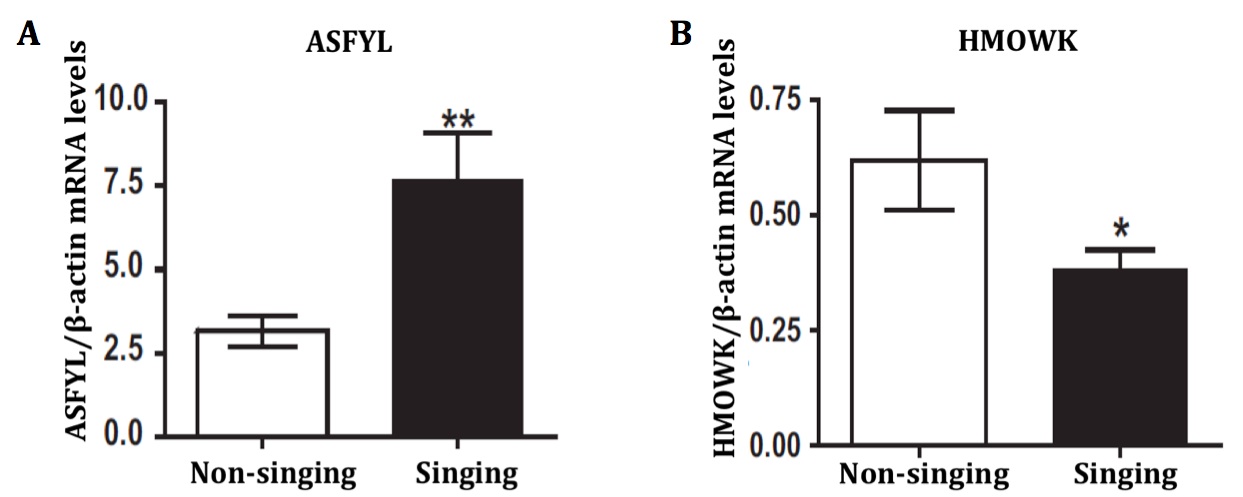
Figure 3 (Click to Enlarge). RT-PCR validation of ASFYL and HMWOK expression. A: ASFYL, identified as hypomethylated in whole blood cells from pups, is expressed at significantly higher levels in those born to singing parents compared to non-singing. B: HMWOK, identified as hypermethylated in whole blood cells from pups, is expressed at significantly lower levels in those born to singing parents as compared with non-singing. Each result is normalized to a housekeeping gene (β-actin) to represent relative levels. Data are means ± SE (n = 6 subjects/group); *P < 0.05; **P < 0.005.
Wookiee duet song ritual enhances force-sensitivity in offspring.
A hallmark of Force aptitude is an elevated midi-chlorian level in cells. Hence, we quantified midi-chlorian DNA in blood samples from pups to determine whether prenatal exposure to duet songs can increase force-sensitivity. We used a multiplex pPCR assay as previously described (Phillips, 2014), and compared the midi-chlorian count per cell among the two groups, singing versus non singing parents. Statistical analysis (Wilcoxon signed-rank test) of mcDNA content revealed that pups born from singing parents had a significantly greater (P = 0.036) number of midi-chlorians per cell that those born to non-singing parents (Fig 4). To test the potential influence of age, an additional statistical analysis was performed with ANCOVA and we found that age was not a significant covariate.
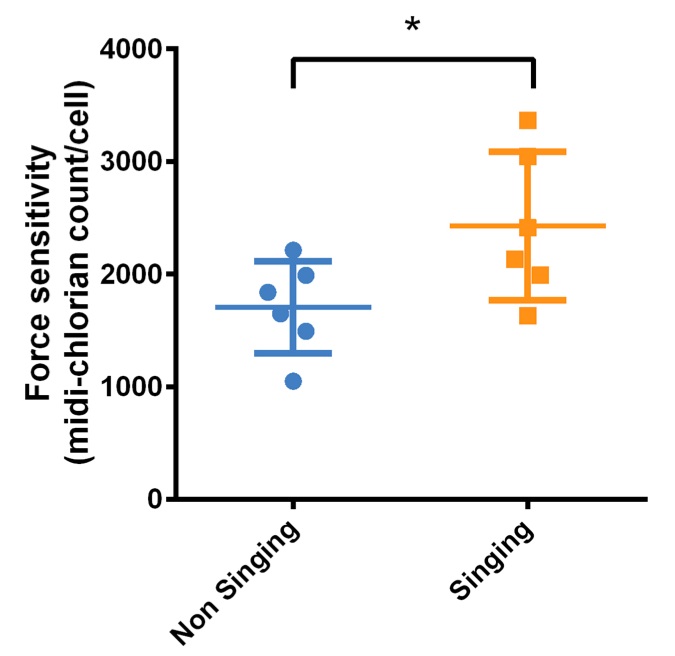
Figure 4 (Click to Enlarge). Force sensitivity is enhanced in pups from parents that produce the duet mating song. Force-sensitivity is represented by midi-chlorian count per cell and was determined by multiplex qPCR for the two groups (n = 6 subjects/group); *P <0.05.
Discussion
Previous attempts in identifying transgenerational effects mating behaviour produces on offspring have been inconclusive. In our study, we found outstanding differences in how genes are regulated epigenetically in pups from singing and non singing parents. The findings from our study can be summarized by four main points: DNA methylation profiles differ depending on whether or not the parents produce the Wookiee mating duet, several force-sensitivity genes are differentially methylated, hypo- and hypermethylation of critical force-sensitivity genes affect the level of gene expression, and pups from singing compared to non singing parents have a greater midi-chlorian content which may be attributed to DNA methylation. Here we provide direct evidence that mating behaviour can produce transgeneration effects on offspring by epigenetic mechanisms.
Our analysis identified five genes involved in the force-sensitivity pathway that are heavily influenced by DNA methylation, UTFL, ASFYL, LSB, NTRP, and HMOWK. This finding supports the notion of a direct link between prenatal care and force-sensitivity of offspring. In particular we found that ASFYL to be hypomethylated and HMOWK to be hypermethylated in pups from singing parents relative to those from non singing parents and that DNA methylation of these genes altered gene expression. Our data demonstrate that differential methylation in response to parental care can regulate gene expression across generations. Our findings also raise new questions such as whether maternal or parental behaviour produce the same effect on offspring, when during development are epigenetic modifications established, and if these changes can be reversed later in life.
It is accepted that midi-chlorians are ancient, symbiotic microbes that have established permanent residency inside the cells of every living form. When midi-chlorians are present in elevated numbers, they confer to the host the ability to detect the Force. A typical human has an average of 2,400 midi-chlorians per cell, while a Wookiee may have anywhere from 1,600-1,800. Our results show that prenatal exposure to Wookiee duet songs can increase the midi-chlorians content, to a level that is comparable to humans. Given that the duet singing custom is rapidly declining in Wookiees, efforts should be made to preserve the culture and to educate Wookiees of its positive benefits to offspring.
Literature Cited
Andreu, A. L., Martinez, R., Marti, R., & García-Arumi, E. (2009). Quantification of mitochondrial DNA copy number: Pre-analytical factors. Mitochondrion , 242-246.
Carvalho, R. H., Haberle, V., Hou, J., van Gent, T., Thongjuea, S., van IJcken, W., et al. (2012). Genome-wide DNA methylation profiling of non-small cell lung carcinomas. Epigenetics & Chromatin , 1-18.
Geissmann, T. (1999). Duet songs of the Siamang, Hylobates syndactylus: II. Testing the pair-bonding hypothesis during a partner exchange. Behaviour , 1005-1039.
Gupta, R., Nagarajan, A., & Wajapeyee, N. (2010). Advances in genome-wide DNA methylation analysis. BioTechniques , 49 (4), iii-xi.
Haimoff, E. H. (1981). Video Analysis of Siamang (Hylobates syndactylus) songs. Behaviour , 128-151.
Khor, G. H., Froemming, G. R., Zain, R. B., Abraham, M. T., Tan, S. K., Tan, A. C., et al. (2013). DNA Methylation Profiling Revealed Promoter Hypermethylation-induced Silencing of p16, DDAH2 and DUSPI in Primary Oral Squamous Cell Carcinoma. Int J Med Sci , 1727-1739.
Organa, L., Blue, D., & Naberrie, P. (2002). Sentient Genetics Omnibus: STAR gene expression and hybridization array data repository. Nucleic Acids Res , 207-210.
Phillips, N. R., Sprouse, M. L., & Roby, R. K. (2014). Simultaneous quantification of mitochondrial DNA copy number and deletion ratio: A multiplex real-time PCR assay. Scientific Reports , 1-7.
Rager, J. E., Bauer, R. N., Müller, L. L., Smeester, L., Carson, J. L., Brighton, L. E., et al. (2013). DNA methylation in nasal epithelial cells from smokers: identification of ULBP3-related effects. Am J Physiol Lung Cell Mol Physiol , L432-L438.
Russell, G., & Rocheleau, G. (2014). Genetic basis for force-sensitivity in Wookies. Annals of Praetachoral Mechanics , 1, 22-28.
Skywalker, A., Fett, J., Marek, G., Offee, B., & Brood, M. (2046). JEDI: Intergalactical Encyclopedia of Genes and Genomes. Nucleic Acids Res , 27-30.
Weaver, I. C., Cervoni, N., Champagne, F. A., D’Alessio, A. C., Sharma, S., Seckl, J. R., et al. (2004). Epigenetic programming by maternal behavior. Nature Neuroscience , 7 (8), 847-854.