DNA FINGERPRINTING IN THE STANDARDIZATION OF HERBS AND NUTRACEUTICALS
(August 2004)
Deoxyribonucleic acid (DNA) is the fundamental building component of all living cells. Our characteristics, traits and physical features are determined by the specific arrangement of DNA base-pair sequences in the cell. It is this distinct arrangement of adenine, guanine, thymine and cytosine (called DNA nucleotides) that regulates the production of specific proteins and enzymes via the Central Dogma Theory [1]. In a living system, this DNA arrangement is uniform throughout the organism, irrespective of the organ. If the DNA from the hair, organs or any body fluid such as blood, saliva or semen, of a particular organism were analyzed, the result would be similar profiles from each. It is this specific 3-D arrangement of DNA that confers upon us our uniqueness in this world. DNA forms the basic genotype (genetic identity) of an organism, which in turn determines the phenotype (physical features) of the organism.
Based on the specificity of the genotype of a system, a particular DNA profile can be ascribed to a particular organism. This profile is as unique as a fingerprint; it is specific to that individual. In the last century, examination of DNA sequences have been used to identify and indict suspects in criminal investigations. Recently, DNA fingerprinting has also been used as a legal tool to determine parentage. Used in combination with forensic and medical evidence, DNA fingerprinting has increased the confidence with which criminals are convicted in court.
This concept of fingerprinting has been increasingly applied in the past few decades to determine the ancestry of plants, animals and other microorganisms. Genotypic characterization of plant species and strains is useful as most plants, though belonging to the same genus and species, may show considerable variation between strains. A good example of this is the fraudulent adulteration of Chianti wines with inferior quality grapes 1.This is also the case with medicinal plants, where the amounts of active chemicals may vary from plant to plant. Herbal drugs are consumed in most developed nations in the form of ethno-therapeutics, nutraceuticals, or are used as the primary source of medicinal compounds or their intermediates. A few plant products commercially available in Canada are from Echinacea purpurea, Panax ginseng, Ginko biloba, Hypericum perforatum, and a host of others. Their commercial value in North America alone spans over $100 million, indicating the volume of use. The varying drug content of different species of herbal plants has been a problem in the production of standardized herbal medicines, where a particular plant from a region can be linked to a specific drug content and thus have a therapeutic value assigned to it, even though similar plants from another region may not share the same levels of the drug. Factors such as soil, climate and adaptability dictate the viability of a particular species and subsequently its drug content. In such cases, there are observed variations in the genetic composition of the plant, in addition to varying amounts of the active drug compound. When used commercially, two factors affect the final drug quality:
(1) The variability with respect to strain-specific drug content;
(2) The potential adulteration of plant drugs with extracts from plants that have lower drug content.
Such discrepancies are very difficult to detect using conventional methods of morphology and microscopy. Cinchona bark, from which quinine is obtained, is one case where the DNA fingerprinting technique could be useful. The bark of Cinchona grown in the plains contains quinine, which is therapeutically active. The same species of tree grown on hilltops and slopes looks morphologically similar but has no active quinine.
In the last two decades, an effective tool to resolve problems in standardization of herbal drugs was devised. Using chemical fingerprinting, plants can be demarcated on the basis of their species, strain and geographical origin. Using chromatographic techniques like High Performance Thin-Layer Chromatography (HPTLC) and High Performance Liquid Chromatography (HPLC), a profile of their various chemical constituents is obtained. This is called chemoprofiling. Chemical constituents are isolated based on their affinities for particular organic solvents in an increasing order of polarity. They are resolved using suitable colouring reagents, resulting in characteristic patterns. The compound specific to that species (sterol, terpenoid, alkaloid, etc.) is characterized as a chemical marker. The medicinal utility of a particular plant species is related to the quantity of that marker compound present. Current editions of herbal pharmacopoeia lay a strong emphasis on the need for chemoprofiling of crude drugs and their subsequent standardization with respect to their composition. A recent offshoot of this method is the use of biomarkers. When using these, the chemical marker compound possesses an intrinsic biological activity. Biomarkers have gained currency primarily in European nations and in countries like China and India, which have a long history of using herbal medicines. The European Scientific Cooperative on Phytotherapy (ESCOP), has clearly specified the requirement of the standardization of phyto-pharmaceuticals on the basis of biomarkers that are unique to that species. Some examples of this are ginsengosides for ginseng and hypericin for St. John’s wort.
Chemical fingerprinting of plants, though possibly informative, does have some pitfalls, which preclude its use as an absolute indicator of the chemical characteristics of plants. In order for a chemical compound to be used as a marker, it must be unique to a particular species of plant. Not all plants contain a unique chemical compound. Even if there is a unique marker, it may not be biologically active. There is a significant overlap of many molecules, especially phenolics and sterols. The main cause of the failure of chemoprofiling is the presence of in-process artifacts, which tend to confound the findings of chemical fingerprinting. Additional techniques are required to profile natural drugs, particularly when profiling the genotypic differences.
Additional motivation for using DNA fingerprinting on commercial herbal drugs is the availability of intact genomic DNA from plant samples after they are processed. Adulterants can be distinguished even in processed samples, enabling the authentication of the drug [2]. Studies have reported the genotyping of several medicinal plants, and have made available their DNA fingerprints. However these results should be taken with a grain of salt as the plants are often sourced from a variety of locations through the world.
Fingerprinting of DNA is dictated by several factors; sequence or restriction site data, taxonomic level of study, the level at which the study is being done (species, genera, etc.), robustness and reproducibility of the method, effectiveness in terms of cost and time, and availability of DNA.
Polymerase Chain Reaction (PCR)
Few techniques in molecular biology have received so much attention and popular acceptance as PCR. Invented by Kary Mullis in 1983, PCR is a method used to generate billions of copies of genomic DNA within a very short time. This amplification is useful in criminal cases where there are miniscule amounts of DNA available. Today PCR finds application in almost all aspects of biomedicine. PCR has been used for the detection of many pathogenic organisms, from bacteria to viruses.
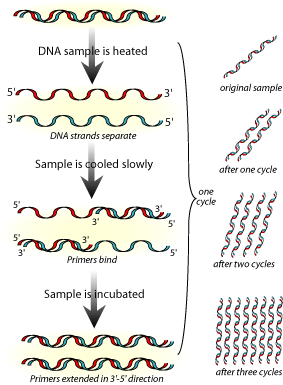
Techniques used in DNA Fingerprinting
1. Microsatellites are simple sequence repeats (SSRs), 1 to 6 nucleotides in length, which show a high degree of polymorphism. Specific microsatellites can be isolated using hybridized probes followed by their sequencing. Like any DNA fragment, SSRs can be detected by specific dyes or by radiolabelling using gel electrophoresis. The advantage of using SSRs as molecular markers is the extent of polymorphism shown, which enables the detection of differences at multiple loci between strains [3].Coupled with chemical and morphological data, we can identify the plant species or strain of interest. The main advantage of using SSRs for fingerprinting is that small amounts of DNA are required compared to the restriction fragment length polymorphisms (RFLP) method. This is due to the large amounts of SSRs present in any genome. Further, assays involving SSRs are more robust than random amplified polymorphic DNA (RAPDs), making them up to seven times more efficient. A drawback to using SSRs is the need to develop separate SSR primer sets for each species. The latest research suggests that SSRs will be involved in new methods of detection of alterations of specific sequences in the DNA.
2. Restriction fragment length polymorphisms are unequal lengths of DNA fragments obtained by cutting Variable Number of Tandem Repeat (VNTRs) sequences up to 30 sequences long with restriction enzymes at specific sites. VNTRs vary between plant species, as do the number and location of restriction enzyme-recognition sites. On an agarose gel, RFLPs can be visualized using radiolabeled complementary DNA sequences. There is no need for PCR amplification of DNA in this method. A routine southern blot experiment is used instead. Normally, RFLPs are used to identify the origins of a particular plant species, setting the stage for mapping its evolution. There are some problems with the RFLP method of DNA fingerprinting. First, the results do not specifically indicate the chance of a match between two organisms. Secondly, the process involves a lot of money and labor, which not many laboratories can afford. Finally, unlike the microsatellites, a few loci in the assay must suffice.
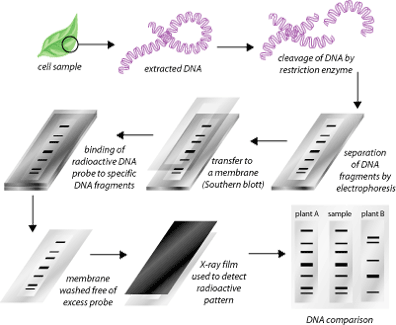
3. Amplified fragment length polymorphism (AFLP) is a PCR-based derivative method of RFLP in which sequences are selectively amplified using primers. It is a reliable and efficient method of detecting molecular markers. DNA is cut with two restriction enzymes to generate specific sequences, which are then amplified suitably. The mere addition or deletion of bases at the 3′ end determines the selectivity and complexity of the amplification 4. By using AFLP, it is possible to evaluate more loci than with RFLP or RAPD. AFLP is also capable of determining a large number of polymorphisms. Similar to SSRs, AFLP-based assays are cost-effective and can be automated.
4. Random amplified polymorphic DNA is one of the most commonly used primary assays for screening the differences in DNA sequences of two species of plants. RAPD consists of fishing for the sequence using random amplification. Here, plant genomic DNA is cut and amplified using short single primers at low annealing temperatures, resulting in amplification at multiple loci. By running a 2-dimensional electrophoresis gel, it is possible to determine the change in sequence pattern by superimposing the 2 gels. Once the band of interest is identified, the gel is cut, and the DNA is isolated and sequenced. Using this target, DNA from other cultivars can be assessed using other techniques such as AFLP or SSRs. It is also more cost effective than RFLPs. RAPDs lack specificity, however, due to low annealing temperatures and easier reaction conditions.
5. Other Methods include the use of single nucleotide polymorphs (SNPs) DNA amplification fingerprinting (DAF) and their offshoots. Although these techniques vary slightly from each other, they operate on the same principle.
Conclusion
DNA fingerprinting, apart from identifying alterations in the genotypes of plant species, is also used for the betterment of drug-yield by tissue culturing. DNA of interest can be stored as germplasm, which is then used for future cultivation. In addition, germplasm can be used for the conservation of selected plant species, which are endangered such as Rauwolfia serpentina (Snake Root). DNA fingerprinting of herbal drugs, though still in its early years, seems to be a promising tool for the authentication of medicinal plant species and for ensuring better quality herbs and nutraceuticals.
Glossary
1. Central Dogma Theory = the fundamental theory of molecular biology that genetic information flows from DNA to RNA to proteins.
2. Nutraceuticals = scientifically designed supplements derived from plants that have proven medical benefits
3. Sterol = a group of steroid alcohols derived from plants or animals
4. Turpenoid = a turpine with an oxygen-containing group.
5. Alkaloid = a nitrogen-containing organic compound, isolated from plants
6. Ginsengoside = active component of the herbal supplement ginseng – has cardiotonic effects, acts on the Central Nervous System..
7. Hypericin = Active ingredient of St.John’s Wort, an herbal supplement used to treat depression, AIDS, and cancer.
8. Phenolic = A compound based on a 6-carbon ring with alternating single and double bonds
9. Cultivar = A “cultivated variety” of plants that are different from normal plants of their species, which retain their differences when they reproduce.
10. Germplasm = Plant materials (germ cells or seeds) that serve as a reservoir of genes for research.
References
1. Breithaupt, H. (2003) Back to the roots EMBO Rep 4(1): 10-12.
2. Mihalov, J. J., Marderosian, A. D., and Pierce, J. C. (2000) DNA identification of commercial ginseng samples J Agric Food Chem 48(8): 3744-3752.
3. Henry, R J. (2001) Plant Genotyping: The DNA fingerprinting of Plants, CABI Publishing, New York.
4. Ha, W. Y., Shaw, P. C., Liu, J., Yau, F. C., and Wang, J. (2002) Authentication of Panax ginseng and Panax quinquefolius using amplified fragment length polymorphism (AFLP) and directed amplification of minisatellite region DNA (DAMD) J Agric Food Chem 50(7): 1871-1875.
(Art by Jen Philpot)