TARGETING YOUR DNA WITH THE CRE/LOX SYSTEM
(August 2004)
It has been 15 years now that the Cre/lox system has been used as a way to artificially control gene expression. If your radar hasn’t picked it up yet, you’re missing out on a clever way to move pieces of DNA around in a cell. Over the years, this system has allowed researchers to create a variety of genetically modified animals and plants with the gene of their choice being externally regulated [1]. This has contributed to our understanding of how individual genes and proteins work.
How it works
The system begins with the cre gene, short for cyclization recombination, which encodes a site-specific DNA recombinase logically named Cre [1,2]. A site-specific DNA recombinase means that the Cre protein can recombine DNA when it locates specific sites in a DNA molecule (see Figure 1). These sites are known as loxP (locus of X-over P1) sequences, which are 34 base pairs long and magnets for the Cre to recombine the DNA surrounding them [2].
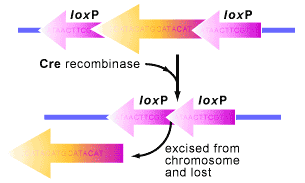
When cells that have loxP sites in their genome also express Cre, the protein springs into action, catalyzing a reciprocal recombination event between the loxP sites (see Figure 1). What does this mean? Well…the double stranded DNA is cut at both loxP sites by the Cre protein and then ligated (glued) back together. As a result, the DNA in between the loxP sites is excised and subsequently degraded. It is a quick and efficient process.
Why Cre/loxP excision of DNA is so useful
The loxP sequence originally comes from the P1 bacteriophage, which is a bacterial virus that, quite reasonably, contains DNA that is not found in animals or plants1. Since the loxP sequences are also 34 base pairs long there is virtually no chance that you would randomly find them in a genome. Therefore, loxP sequences can be artificially inserted into animals or plants and used for the precise excision of DNA, without worrying about cutting other parts of an organism’s genome.
Regulating a Gene Using the Cre/loxP System
For a gene to produce a protein it requires a ‘promoter.’ This is a section of DNA in front of the gene that functions to recruit the cellular machinery that will initiate the multi-step process of protein production (called gene expression). How the promoter functions to do this can vary, from always recruiting cellular machinery and thus always being ‘on’, to only doing this in specific tissues or cell types, or being inducible and thus only functioning in the presence of a specific factor or condition. Each type will dictate the amount of protein a gene can produce and thus ultimately control aspects of its function. For this reason, a promoter is considered a ‘regulator’ of gene function.
An artificial type of this regulation can be achieved with the Cre/lox system [1,3-4]. To do this you need to create a transgenic organism with an always ‘on’, or ubiquitous, promoter that is attached to and ready to regulate the gene you would like to control. The trick is: in between the promoter and the gene, a ‘stop’ sequence surrounded with loxP sites is inserted (see Figure 2). The stop sequence is a short sequence with several transcriptional stop codons (terminators) that will prevent the gene from producing a protein.
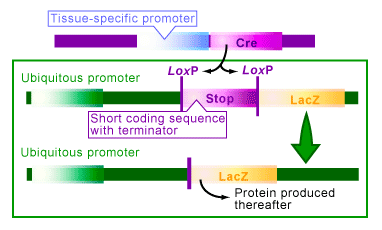
Take the example in Figure 2, where you are trying to control the gene lacZ. Transgenic organisms with a ‘stop’ sequence between the promoter and gene cannot express the LacZ protein. When Cre is present in the cells of this organism, it catalyzes recombination between the loxP sites, thereby deleting the ‘stop’ sequence. With the stop sequence deleted the organism would express LacZ, or whichever other gene that was being studied in its place. The conditions in which the Cre is present thus regulated the expression of the gene.
The Cre/lox System in Action
An example of transgenic mice provides an example of how the Cre/lox system can be used. In this case we would like to produce a mouse that no longer has a target gene in only one cell type. The Cre/lox system provides a method to do this [4] (see Figure 3):
1. Transgenic mice containing a gene surrounded by loxP sites are mated with transgenic mice that have the cre gene expressing only in one cell type.
2. The resulting mice with both the cre gene and the loxP-flanked gene.
3. In tissues with no cre gene the target gene with be present and function normally.
4. In a cell where Cre is expressed, the target gene of interest will be deleted.
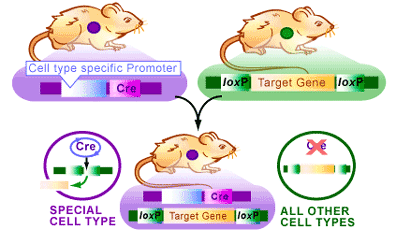
Therefore, if the cre gene is bound to a promoter that only allows Cre production in neuronal cells, the target gene will be deleted only in those cells. This method allows researchers to isolate the effects of genes in specific tissues, thereby providing very specific analysis of gene function. Since the Cre/lox system has been used extensively over the last fifteen years, there are now numerous animal, plant and bacterial stocks that already contain the cre gene driven by ubiquitous or tissue-specific promoters. These established lines provide a quick method for breeding experiments like those described above.
Advantages and Disadvantages
The Cre/lox system has the advantage of working in almost any type of cell. This versatility has led to its application in many types of experiments, such as, labelling neuronal cells in the brain, differentiating them from surrounding glial cells and looking at the properties of promoters [5]. A more powerful approach to controlling gene expression has also been developed by combining the Cre/lox and the tetracycline-regulated transcriptional systems [6]. In this combination system expression of Cre occurs only when the drug tetracycline is administered, allowing researchers to start and stop expression of the gene at any time. The two primary disadvantages of the Cre/lox system are that not all tissue specific promoters are perfectly specific [4]. Basal levels of expression in other cell types can sometimes cause unintended gene expression. In addition, the establishment of transgenic systems with inserted genes requires a significant amount of time and money.
A Final Thought
The Cre/lox system is an invaluable tool for molecular biology. By establishing tissue specific expression, it allows the isolation of individual genes and their functions. Controlling genes through the Cre/lox system is comparable to controlling a toy car. You can select the area where the gene will be expressed (direction) and control the level (speed) at which the gene will be expressed.
Additional Reading
1. Latchman DS. (2002). Gene Regulation: A Eukaryotic Perspective. Cheltenham: Nelson Thornes. 323p.
2. Perkins AS. (2002). Functional Genomics in the Mouse. Functional & Integrative Genomics 2(3): 81-91.
3. Nagy A, Mar L. (2001). Creation and use of a Cre recombinase transgenic database. Methods in Molecular Biology 158: 95-106.
References
1. Kuhn R, Torres RM. (2002). Cre/loxP Recombination System and Gene Targeting. Methods Mol Biol 180: 175-204.
2. Ghosh K, Van Duyne GD. (2002). Cre-loxP Biochemistry. Methods 28(3): 374-83.
3. Le Y, Sauer B. (2000). Conditional Gene Knockout using cre Recombinase. Methods Mol Biol 136: 477-85.
4. Brian Sauer. (1998). Inducible Gene Targeting in Mice Using the Cre/lox System. Methods 14(4): 381-92.
5. Malatesta P, et. al. (2003). Neuronal or glial progeny: regional differences in radial glia fate. Neuron 37(5): 751-64.
6. Utomo AR, et. al. (1999). Temporal, spatial, and cell type-specific control of Cre-mediated DNA recombination in transgenic mice. Nature Biotechnology 17(11): 1091-6.
(Art by Jane Wang – note that high res versions of image files available here)