EXPRESSION OF MaGc77 IS A SPECIFIC AND SENSITIVE MARKER FOR THE PATRONUS (Pt) CHROMOSOME (MGR-ABL1) TRANSLOCATION IN INHERITED MAGICAL ABILITIES
Annals of Praetachoral Mechanics (2016). Vol 2. Advanced online publication. download pdf
ABSTRACT
Magical abilities have been estimated to occur in about 17% of the human population. However, the cause of discrepancy between wizards/witches and muggles (non-magical humans) remains largely unknown. More surprisingly, the presence of squibs (individuals with no magical abilities despite a magical heritage) and muggle-born wizards and witches indicates a genetic component to magical abilities, which has yet to be understood. Recently, the MaGc77 protein was described as playing a potential role in the conference of magical abilities in individuals. Here we describe the underlying genetic event resulting in overexpression of MaGc77 in magical individuals. Next generation sequencing technologies unveil the MGR-ABL1 translocation event to be recurrent in magical individuals. Furthermore, our study demonstrates the overexpression of MaGc77 transcription factor in magical individuals. Isolation and transfusion of recombinant MaGc77 could be a promising strategy for induction of magical abilities in muggles.
Keywords: MGR-ABL1, MaGc77, Patronus, Magical Ability, Muggle, Squib
Introduction
MaGc77-positive individuals (“magical people”) have been the object of extensive studies to define, at a molecular and cellular level, the underlying mechanisms which may lead to the generation of this protein as it is not encoded by a single gene. In addition, the complex pathways, whereby this protein may confer magical abilities, are only partially understood. Although the expression of the hybrid MaGc77 protein is presumed to occur early in childhood or adolescence, and entails elevated tyrosine kinase activity, the intimate role of the MaGc77 protein in magical transformation has not yet been completely elucidated. This protein was originally described in individuals with acute magical abilities, but has since been shown to be present in almost all individuals with magical abilities: consequently, it is now considered the hallmark of this supernatural ability (Rowling et al., 1984). However, unlike other protein expression profiles (e.g. Kedavra86, Cru and MgiC100) that are specifically associated with particular types of magical abilities (McGonagall, 1990), the MaGc77 protein is not restricted to pure bloods, but can also be found in a significant proportion of squibs and wizards/witches of non-magical heritage (muggle-born) (McGonagall, 1991).
We had previously described a relatively weak and rather common form of magic in children that we called ‘Underage Magic Without Horcruxes and Teleportation’ (UM-WHAT) (Mottok et. al., 1995), which is associated with the MGR gene. Here, we identify magical individuals carrying a previously undescribed type of MGR-ABL1 gene fusion that shows a novel position for a breakpoint in the MGR gene far downstream from other breakpoints described previously (Hagrid et al., 1996). This has led to the hypothesis that the structure of this gene and, in particular, the location of the breakpoint in MGR correlates with the magical phenotype (Hagrid et al., 1996).
While it is difficult to experimentally create magical conditions in the lab, we have managed to obtain biological samples from magical individuals to create a cohort with MGR-ABL1 fusions, which actively express the MaGc77 nuclear transcription factor. The precise function of this protein is yet to be elucidated. We used a wide variety of high-throughput sequencing techniques as well as bioinformatic tools to uncover the underlying genetic mechanism behind magical capabilities. Here, we show the genetic translocation event which leads to the fusion gene and uncovers a pro-magic role of MaGc77 in the conference of magical abilities in individuals. We postulate that the isolation and subsequent transfusion of MaGc77 into muggles should confer them with magical powers.
Methods
Human samples.
Peripheral blood was obtained from three volunteers (HP, RW, HG; discovery cohort) and used for BAC capture and whole transcriptome sequencing, respectively. The extension cohort comprised of tissue specimens from 55 individuals, which were available from the tissue archives of the Centre for Applied Translational Witchcraft and Wizardry. This study abided by the Declaration of Hogsmeade and was approved by the Hogwarts School of Witchcraft and Wizardry Ethics Board (H08-00666).
BAC capture.
To explore novel structural genomic rearrangements and breakpoint anatomy at basepair resolution in individuals with extraordinary magical capabilities, we selected three individuals to perform targeted BAC capture high-throughput sequencing. DNA was extracted from peripheral blood samples using the Qiagen QIAamp DNA Blood Maxi Kit as per the manufacturer’s protocol and 2.2 μg were used for library construction. 77 BACs tiling a total of 7.77 Mb across 20 genomic regions on 10 different chromosomes known to be recurrently affected by rearrangements in intellectually gifted people were selected to enrich the libraries. Briefly, BAC clone DNA was extracted and the identities were confirmed by BAC-end Sanger sequencing using T7 and SP6 primers and DNA fingerprinting. Validated DNAs were pooled, sonicated and products in the 75-200 bp range were excised from an 8.8% PAGE gel and quantified. 222 ng of the size-selected pooled DNA was end-repaired and phosphorylated for ‘A-tailing’ and adapter ligation. Adapter-ligated products were enriched using 8 cycles of PCR. Gel-purified DNA was in vitro transcribed using T7 RNA Polymerase. Libraries were captured using a 444:1 ratio of BAC-derived probes to library DNA. Products underwent 11 cycles of PCR post-capture prior to sequencing. 75 bp paired-end sequencing was performed on an Illumina Hi-Seq2000 and analysed using the nFuse and deStruct algorithms to generate and cross-corroborate rearrangement predictions as previously published (Kupferfeld, 2011; Simon and Garfunkel, 2012). Predictions were filtered on the basis of a validation probability metric (>0.75); a score of uniqueness among libraries, quality and number of spanning and split read support, and alignment relative to the capture space. Subsequently, these events were BLAT-assessed and subjected to validation using Sanger sequencing.
Whole transcriptome sequencing (RNAseq).
RNA from the same peripheral blood samples as described in the previous section was extracted using the RNeasy Mini Kit (Quiagen). RNAseq was performed as previously described (Mottok, 1995). In brief, double stranded cDNA was synthesized from polyadenylated RNA and sheared. The 180-200bp fraction was isolated and amplified with 11 cycles of PCR using the Illumina Genome Analyzer paired end library protocol. The resulting RNAseq libraries were then sequenced on an Illumina Genome Analyzer and aligned to hg007 using GSNAP55. Gene fusions were predicted using deFuse and reported based on the following criteria: (1) altsplice = N, (2) cDNA_breakseqs_percident < 0.5, (3) breakseqs_estislands_percident < 0.5, (4) genome_breakseqs_percident < 0.5, (5) break_adj_entropy_min > 0.5, (6) probability > 0.5, (7) one of the breakpoints was in a coding region, and (8) gene fusion partners are not adjacent.
Validation of predicted alterations by PCR and Sanger sequencing.
All predictions resulting from BAC capture and RNASeq were subjected to validation using Sanger Sequencing (3130 Genetic Analyzer, Applied Biosystems, Foster City, CA or outsourced to Genius Wizard Inc., London, ON). Prior to Sanger sequencing the genomic region or the predicted transcript was amplified from DNA or cDNA by standard PCR. Sequence analysis and assembly was performed using MagicClone manager software. Genomic coordinates are given according to hg007.
Flow cytometry.
Flow cytometry was used to quantify MaGc77 and ABL1 expression in peripheral blood cells obtained from the three individuals of our discovery cohort. Cells were first fixed in 0.01% formaldehyde for 10 minutes and permeabilized by adding 1 ml methanol to each sample. Cells were then placed at -20°C for 10 minutes and were subsequently washed twice in cold PBS containing 1% FBS. 0.5 million cells were then incubated for 30 minutes on ice with PerCP/Cy5.5 anti-human ABL1 (Biolegend, San Diego, CA, 0.25 ug/test) and FITC anti-human MaGc77 (kind gift from Dr. D. Kupferfeld). After washing and re-suspension, samples were run on a FACSCalibur (BD Biosciences). All analyses were done using FlowJo software (version 10.0.7r2, FlowJo, LLC, Ashland, OR).
Fluorescence in-situ hybridization.
The recurrence of the MGR-ABL1 rearrangement was assessed using the customized spectrum green and orange bacterial artificial chromosome probes covering the MGR gene locus and the same tissue microarray as for immunohistochemistry (see below for detailed information). 200 interphase nuclei from at least four x400 fields of view were independently evaluated by two investigators with an additional 100 interphase nuclei assessed by an independent analyzer in the event of discordant results. Images were captured at room temperature using an automated Zeiss Imager.Z2000 fluorescence microscope, Metafer software and Metafer CoolCube 1 camera (MetaSystems). Cases in which > 5% of the nuclei housed a split of the red and green signals or individual loss of either the red or green signal were categorised as break-apart positive.
Tissue microarray construction and immunohistochemistry (IHC).
All tissue samples underwent a centralized review in order to select the area of the paraffin block for tissue microarray (TMA) construction. Duplicate 1.5 mm cores from 55 cases were assembled on one TMA. Following TMA construction 4μm slides were cut and immunohistochemical staining was performed using routine protocols for automated procedures on a Benchmark XT platform (Adventura, KU). Specifically, sections were stained with an Common Welsh Green dragon anti-MaGc77 antibody (1:10,000; Nanopore, NN). Protein expression was independently determined and recorded as the percentage of positive cells in 10% increments by two magical pathologists (GK and AM).
Results
A gene fusion event occurs between MGR and ABL1.
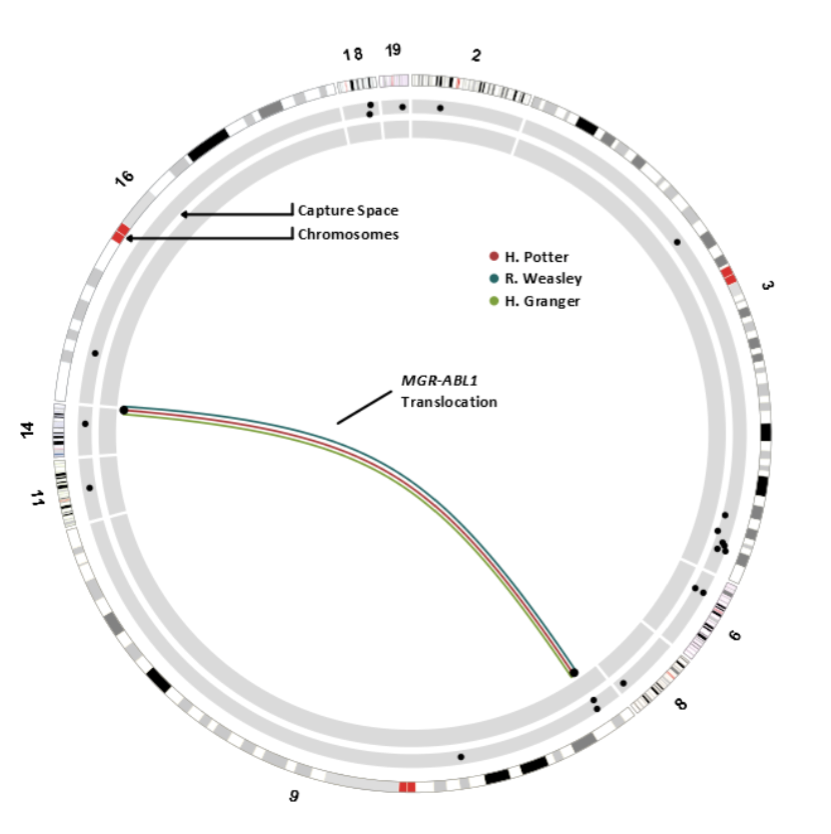
BAC capture sequencing yielded a mean coverage depth of 99X across the capture space in the three enriched libraries; 66.6% of reads were non-PCR duplicates that aligned to hg007/GRCh0815. Following stringent filtering, a list of 13 potential genomic rearrangements in the three libraries was generated. All these predictions were validated using Sanger sequencing (data not shown). Since the translocation involving MGR on chromosome 14 and ABL1 on chromosome 9 was the only genomic event which could be recurrently detected across all three libraries (Figure 1), we decided to focus on the characterization of this aberration.
(CLICK ON IMAGE TO ENLARGE) Figure 1. Circos plot depicting the 20 regions of bacterial artificial chromosome capture space spanning 7.77 Mb and the recurrent structural genomic event in three libraries. The outer-most concentric track denotes the chromosomes and the middle track denotes the capture space (see Table S1 for additional details) with black dots representing the genes covered by the BACs. The adjoining lines depict rearrangement partners. Events are colour-coded to reflect those which overlap between cases. Only chromosomes relevant to the capture space have been included and chromosomes 3, 9 and 14 have been magnified to improve resolution. The regions of the chromosomes that are highlighted in red are the centromeres.
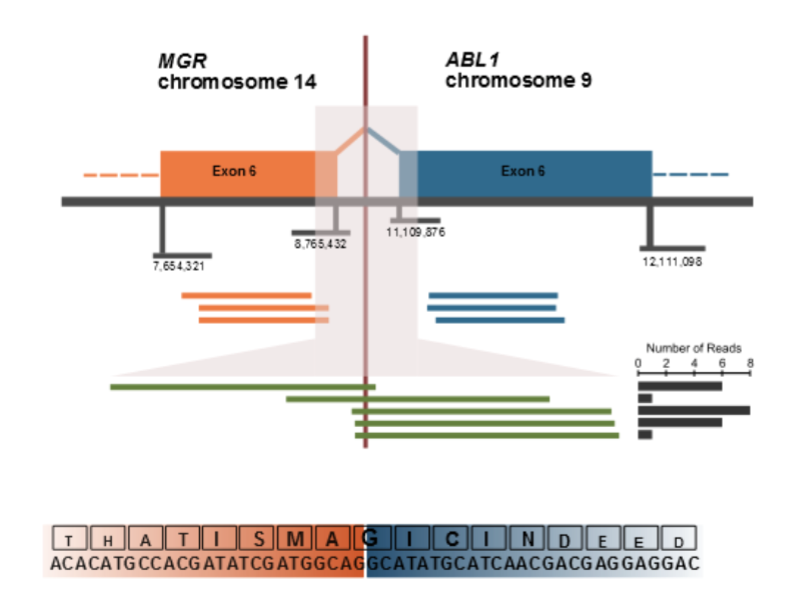
The precise genomic breakpoints for the MGR-ABL1 fusion were identified to be: chr9:g.8,888,888 and chr14:g.10,000,010 and were validated using Sanger sequencing. We interrogated these findings with results obtained from whole transcriptome sequencing. The analyses led to the discovery of an in-frame MGR-ABL1 fusion transcript with the amino terminus of the predicted chimeric protein encoded by the first six exons of MGR and the carboxyl terminus encoded by exons 6 to 11 of ABL1. Figure 2 depicts the precise genomic coordinates, the sequence of the fusion transcript and the effect at the transcript level (Figure 2). Since transcription of this fusion would be driven by the strong MGR promoter, expression of the fusion transcript (and the 3’ portion of ABL1) from this allele would be expected to be elevated. Indeed, whole transcriptome sequencing revealed, that >98% of MGR and ABL1 transcripts are derived from the allele harboring the gene fusion, suggesting that this acts as a dominant positive variant.
(CLICK ON IMAGE TO ENLARGE) Figure 2. MGR-ABL1 fusion observed using paired-end whole transcriptome sequencing. The upper panel shows the genomic structure with paired read sequences aligning on either side of the breakpoint (orange and blue lines). Split reads spanning the breakpoint are highlighted in green with the histogram on the right showing the absolute frequency for each of these reads. The lower panel provides the reading frame at the breakpoint junction and the putative translation.
MGR-ABL1 fusion event leads to the increased expression of MaGc77.
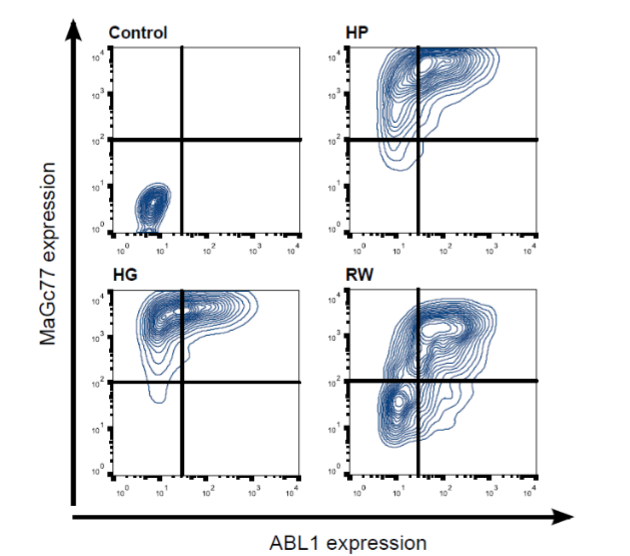
To correlate the fusion event with the overexpression of MaGc77 protein, we assessed ABL1 and MaGc77 protein expression by flow cytometry. Compared to a muggle (with no magical capabilities) expression of both proteins were highly elevated in our discovery cohort (Figure 3). This study was then extended to a cohort of 10 magical individuals, who had donated blood samples in return for butter beer. A similar increased expression of both proteins was seen in 8/10 (80%) of these individuals (data not shown). Although the expression levels of MaGc77 for all individuals was beyond the magically significant threshold (77%), the level of expression of MaGc77 seemed to be directly correlated to the power and incantation wisdom possessed by the individual. The power of the individuals was assessed by a simple test of efficacy of the patronus charm.
(CLICK ON IMAGE TO ENLARGE) Figure 3. Flow cytometry for MaGc77 and ABL1. Density plots for MaGc77 and ABL1 expression are depicted. The control sample is negative for both markers whereas the peripheral blood cells of the three individuals of the discovery cohort show elevated but variable expression of both markers as a result of the MGR-ABL1 fusion.
FISH studies exhibit break apart signals in magical individual.
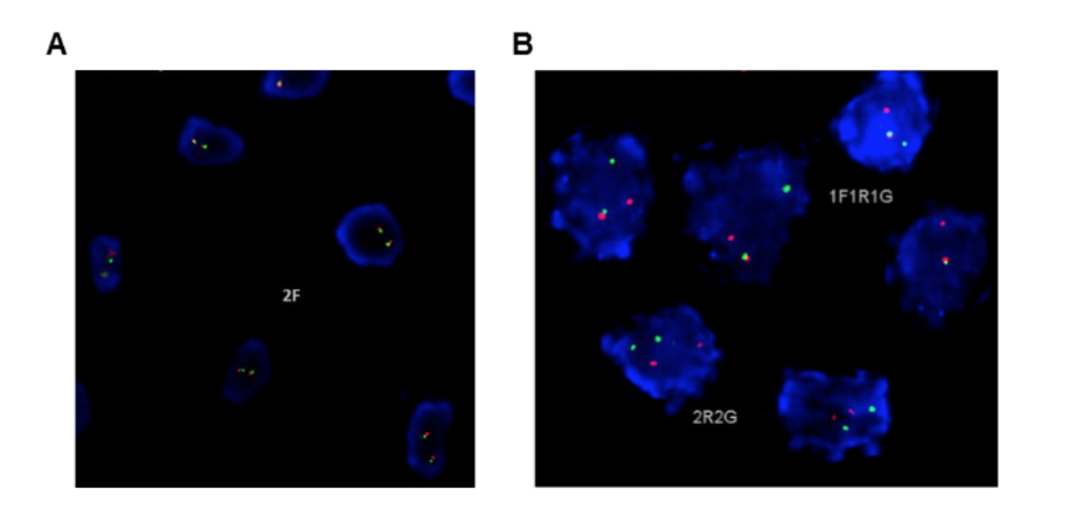
Extending our study of MGR rearrangements in magical individuals, we performed FISH on additional 55 specimens from magical individuals using a previously reported MGR break-apart assay (Figure 4). Of 55 initially assessed specimens, a total of 46 (84%) possessed an interpretable signal pattern. In total, we observed a break-apart signal pattern in 35/46 (76%) of evaluable cases, suggesting that MGR rearrangements are recurrent and may be able to predict learning success and magical powers in at least a subset of these individuals.
(CLICK ON IMAGE TO ENLARGE) Figure 4. Fluorescence in-situ hybridization (FISH). FISH was performed using a break-apart-assay for the MGR gene locus. Representative images of a translocation-negative case (A) with two fusion signals and a rearranged case with a split-signal constellation (B) are shown.
IHC reveals that rearrangements resulting in the MRG-ABL1 fusion are associated with increased MaGc77.
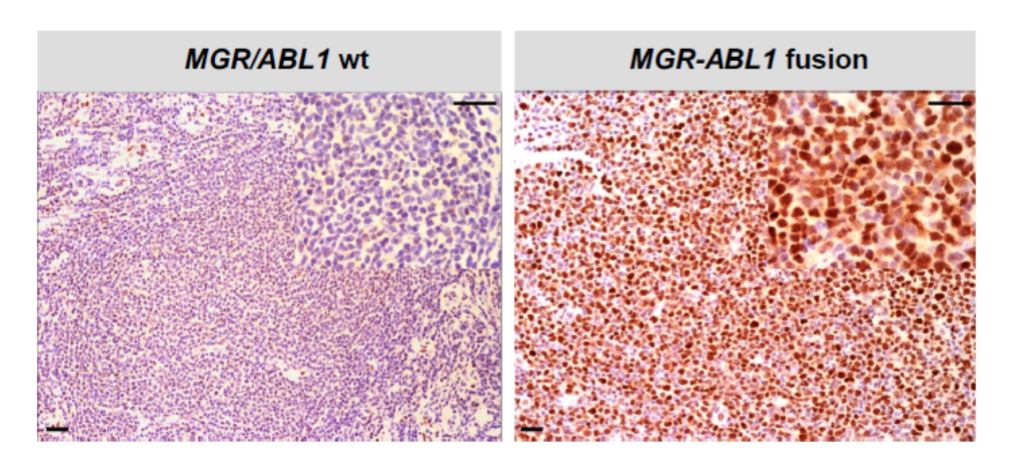
To determine the overall frequency of MaGc77 expression in the Hogwarts School of Witchcraft and Wizardry student cohort, immunohistochemistry was performed on the tissue specimens, sampled on the TMA (Figure 5). Immunohistochemistry was evaluable in 53/55 cases and a moderate to strong staining was observed in 40 of these 53 samples (75%). Correlation with the MGR break-apart status revealed a strong and significant correlation (Spearman correlation coefficient r=0.90, P=0.001) of protein expression and MGR rearrangements. These results clearly demonstrated a correlation between the MGR-ABL1 fusion and the overexpression of MaGc77.
(CLICK ON IMAGE TO ENLARGE) Figure 5. MaGc77 immunohistochemistry staining pattern. Intense nuclear staining for MaGc77 in the sample on the right compared to a control case on the left side. The overexpression of MaGc77 protein informs on the functional consequences of the translocation involving the MGR and ABL1 genes. Representative pictures were taken using a Nikon Eclipse E600 microscope equipped with a Nikon DS-Fi1 camera at room temperature. Original magnification: x100 and x400 for the inserts; black bars: 50 μm.
Discussion
As far as we are aware, this is the first attempt to define the genomic translocation fusion event which leads to the overexpression of MaGc77 and subsequent conference of magical abilities and represents a very novel finding. We have decided to call this the Patronus (Pt) chromosome. The initial analysis of this data confirms that the status of MGR-ABL1 rearrangement and in particular, the resultant increased expression of MaGc77 is a key factor in determining the magical status of an individual. Thus we conclude that this is a genomic alteration which occurs frequently in magical individuals. Furthermore, we believe that this chromosomal event is highly inheritable but can occur in a sporadic manner as well (thus accounting for muggle-born wizards and witches). Subsequent interrogation will however need to be done to determine exact inheritance patterns as well as precise downstream targets of MaGc77.
Our findings demonstrate at base-pair-level resolution, a novel structural genomic rearrangement resulting in a fusion transcript and chimeric protein in people with magic capabilities. We have demonstrated that this rearrangement substantially alters protein expression in primary tissue samples.
This study has overarching implications for both the magical and muggle community. Prior to this study, attempts had been made to confer magical abilities in muggles or squibs by means of magical incantations or education. However, these attempts have been largely unsuccessful. We postulate that with our current findings, it could be possible to isolate and transfuse MaGc77 into individuals lacking magical abilities. While we believe the powers conferred by this means may be temporary, a more permanent solution would have to be developed.
Acknowledgements
The authors would like to thank Harry Potter, Ron Weasley and Hermione Granger, for graciously accepting to part of the discovery cohort. In addition, the authors would also like to acknowledge the contributions of the students of Hogwarts for being part of the study cohort. Finally, the authors would like thank Headmistress Minerva McGonagall for graciously allowing us to carry out our experiments in the old potions dungeons as well as giving us access to prior confidential records.
References
1. Hagrid, R., et al (1996). Classification of magic phenotype associated with the MGR gene. Nature Magic 1232(11), 12-257.
2. McGonagall, M., et al. (1990). Magical proteins and where to find them. Journal of Magical Exploits, 3455(12), 1426-1429.
3. McGonagall, M., et al. (1991). To magic or not to magic: A study of inherited magical abilities in witches and wizards. Journal of Magical Biology, 1(7), 313- 315.
4. Mottok, A, et al. (2012). UM-WHAT is occurring in wizarding children? Nature Magic, 3401(24), 123-137.
5. Kupferfeld, D, et al. (2011). nFuse can predict magical fusions. Nature Magic Technologies 777(33), 99-111.
6. Simon and Garfunkel (2012). The deStruct algorithm is a useful tool to predict novel structural genomic alterations in magical individuals. Magic Science 654(77), 1-12.