IDENTIFICATION OF HUMAN CHROMOSOME γ AND ITS EPIGENETIC CONTROL FOR THE EXPRESSION OF “SUPER” GENES
– – –
New England Journal of Medicine. (March 2013). Vol 328 No 12. DOI: 10.1738/NEJMoal219833 – pdf download
ABSTRACT
BACKGROUND
Superheroes possess exceptional characteristics that far exceed the scope of human capabilities, such as the tremendous strength and pigmentation observed in The Incredible Hulk following his exposure to γ-radiation. Here, we explore the underlying molecular mechanisms for the “superhuman” abilities seen in the Hulk, as compared to semi-superheroes Papa Smurf and Popeye the Sailor. In particular, we examine the role of epigenetic mechanisms, principally DNA methylation, in the genotypic expression of “super” genes and their phenotypic manifestation.
METHODS
Whole Transcriptome Shotgun Sequencing (WTSS) was conducted on The Incredible Hulk, Popeye the Sailor, and Papa Smurf to identify “super” genes. Whole Genome Shotgun Sequencing (WGSS) was subsequently performed on the Hulk, from which chromosome γ was assembled and Bacterial Artificial Chromosomes (BAC) were created. FISH-karyotyping was then employed for chromosome examination before and after treatment with γ-radiation. Methylation assays were conducted on CpG islands upstream of “super” genes to assess gene expression following treatment with activator substances spinach and smurf berries.
RESULTS
Two highly expressed novel genes, Vis and Colos, were identified by WTSS in The Incredible Hulk. Vis was also expressed in Popeye, suggesting that it is associated with superhuman strength, and Colos was also expressed in Papa Smurf, suggesting that it is associated with hyperpigmentation. WGSS revealed that the genes are located on opposite arms of a novel chromosome. This chromosome was confirmed by karyotyping and named chromosome γ. FISH probes revealed that in all cases, save the Hulk, the γ-chromosome was associated with the centromere of the X-chromosome. Treatment of normal cells with γ-radiation caused a dissociation of the γ- from the X-chromosome. In in vitro studies, it was found that concentrated spinach and smurf berries decrease methylation in CpG islands upstream of the Vis and Colos genes, respectively.
CONCLUSION
There exists a novel chromosome, which is somehow intertwined with the centromeric region of the X-chromosome. It contains two “super” genes, for strength and color, which under normal conditions are silenced by upstream CpG island methylation. These genes can be activated by spinach or smurf berry treatment (via CpG demethylation) or by exposure to γ- radiation (via physical separation of chromosome γ from X). This suggests a potential for the discovery of additional “super” genes and chromosomes in association with previously defined chromosomes.
INTRODUCTION
SUPERHEROES POSSESS EXTRAORDINARY OR unusual superhuman powers [1]. It has been proposed that alternative gene regulation or genetic mutations are the root of such exceptional phenotypic abilities [1,2,3]; however, these genotypic abnormalities remain poorly defined. Understanding the molecular mechanisms responsible for expression of “super” genes is pivotal in determining the physiological and cellular features conducive to superhuman abilities.
Epigenetic regulation of gene expression, such as DNA modification, has been shown to direct the transcriptional activity of genes [4-7]. DNA methylation is a common epigenetic marker and plays important roles in the regulation of gene expression, genomic imprinting, X-chromosome inactivation, embryonic development, and cancer5. Methylation of CpG islands upstream of promoter regions by DNA methyl-transferases decreases transcriptional activity of those genes, whereas demethylation increases activity [4,7]. The process of DNA methylation is a dynamic and reversible enzymatic process [8,9].
The Incredible Hulk is a gigantic green human-like antihero possessing incredible strength. He is the transformed alter ego of Dr. Bruce Banner, that is a result of exposure to a “lethal” dose of gamma radiation [10]. Since this initial transformation, Dr. Banner can transform into the Incredible Hulk whenever enraged or endangered [11,12]. The central molecular mechanisms responsible for this dramatic transformation have yet to be determined. Here, we identify a novel chromosome associated with the X-chromosome contain-ing two genes that contribute to the observed Hulk phenotype. We used for comparison two semi-superheroes, Papa Smurf and Popeye the Sailor, who possess similar phenotypes to the Hulk: abnormal color and immense transient strength, respectively.
METHODS
SAMPLE ACQUISITION
Primary B lymphocytes were sourced from cervical lymph node biopsies (performed by A. Y. J.) of four index cases: The Incredible Hulk, Popeye the Sailor, Papa Smurf, and a healthy 42-year-old male human. For the cases of the Hulk and Popeye, two independent biopsies were performed: one during the resting state and (at considerably greater difficulty) one during the peak phase of strength.
CELL LINE GENERATION
B lymphocyte cell lines were generated from all biopsies. B-cells were selected for using CD19 antibody-targeted FACS (BioLegend, cat. 302234) and validated by a hematopathologist (R. D. G.). Cells from Papa Smurf (PS-1), active Hulk (Hulk-1), and active Popeye (PE-1) were found to grow well in 100% heat-inactivated fetal bovine serum (HI-FBS), at 3% CO2 and 42 oC.
In establishing cell lines from the human control (LCL-49), non-active Hulk (NAH-20), and non-active Popeye (NAP-9), transformation with competent Epstein-Barr virus was required. Subsequently, these cell lines were found to grow modestly in 20% HI-FBS RPMI-1640 media (Gibco, cat. 11875-093) containing 1% penicillin/streptomycin, 2 mM L-glutamine, 1 mM sodium pyruvate, and 0.1 mM non-essential amino acid at 5% CO2 and 37 oC. After ~5 passages, cell lines LCL-49, NAH-20 and NAP-9 were found to either no longer to exhibit B-cell characteristics or be competent.
PREPARATION OF NUCLEIC ACIDS
Both DNA and RNA were extracted from all cell lines using Qiagen’s Allprep Mini kit (cat. 80004). cDNA was synthesized using the Invitrogen Super-Script III Reverse Transcriptase kit (cat. 11754-050).
WHOLE-TRANSCRIPTOME SHOTGUN SEQUENCING OF HULK cDNA
WTSS was performed as previously described [12]. Briefly, this involved the synthesis of cDNA from transcript RNA, the sonic shearing of cDNA to produce fragments <300 bp in size and the amplification of products 32,768-fold, by means of the Illumina Genome Analyzer paired-end library protocol. The generated library was sequenced using an Illumina Genome Analyzer IIX.
Reads were aligned to the NCBI build 36.2 hg19. Those reads not aligning to hg19 were binned and aligned using the Cufflink algorithm (from Tophat, version 2.0.2) which is also capable of calculating transcript abundance in addition to assembling reads de novo.
WHOLE-GENOME SHOTGUN SEQUENCING OF HULK γDNA
WGSS was performed as previously described [13]. 1 µg of genomic DNA was shredded and size-selected for sequences <400 bp. Barcode and priming sequences were annealed to the size-selected DNA prior to 10 cycles of amplification observing the Illumina PCR protocol. The Hulk genomic library was run on an Illumina HiSeq 2500 (16 lanes for >2,000 x coverage).
As with the WTSS, reads were aligned to hg19. Non-aligning straight and paired-end reads were analyzed with the assistance of the Wang group for the assembly of chromosome γ [14].
FISH-KARYOTYPIC ANALYSIS
From the WGSS analysis, novel BAC probes were generated, as previously described [15], to include ~100 kbp sequence upstream of Vis and Colos (see Fig. 1). Samples from all cell lines were subsequently M-phase methotrexate-synchronized, ~105 cells were fixed to slides, and probes were hybridized according to the Genetic Pathology Evaluation Centre protocol [16]. Chromosomes were visualized using a Zeiss automated microscope with analysis software (CytoVision) from Ariol.
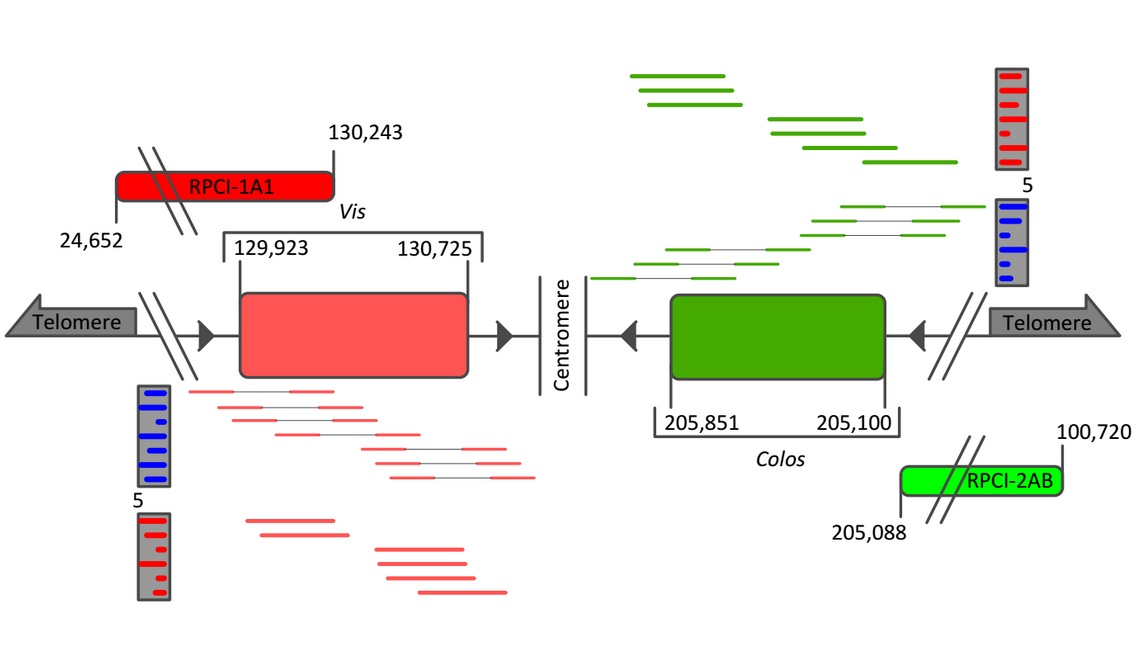
Figure 1: WGSS sequencing the Hulk. Both genes were found to be one exon is size and Colos (751 bp) as presented is relative in size to Vis (802 bp). Approximate paired-end read support is provided in blue (Vis = 27 reads, Colos = 21 reads), while the full length read support is provided in red. Split and full-length reads are not to scale. Newly generated BAC probes (RPCI-1A1 and RPCI-2AB) are mapped relative to Vis and Colos exons.
IN VITRO γ-RADIATION
Cell lines were exposed to gamma-radiation in increments of 50 Gy (10-510 Gy). A Gammacell 220-cobalt-60 device (Atomic Energy of Canada Limited), producing 30 Gy/min, was used to generate γ-radiation [17]. Following this exposure, a FISH-karyotypic analysis was performed, as described above.
METHYLATION ASSAY
Spinach purée was generated by means of dicing and steaming 50 kg of spinach for 5 hours at 60 oC and 10 PSI. The resulting purée was subject to 47 rounds of repeated filtration and evaporation to distill to a concentrate of 2 ml. This protocol was observed to generate 2 ml of smurf berry concentrate, using 7 rounds of repeated filtration and boiling. A co-culture treatment matrix was then devised to expose 2 million cells (enumerated via Invitrogen’s Countess device) from each cell line to spinach and smurf berry concentrate (ranging from 100-2000 µl of concentrate per 10 ml culture). Cells were co-cultured for 1 hour in respective concentrates at ideal growth conditions, after which genomic DNA was harvested as before.
1 μg genomic DNA from each cell line treatment was bisulfate-converted using the EZ DNA Methylation kit from Zymo Research (cat. D5002). In working with Affymetrix, a novel chip was developed for the γ-chromosome to which genomic DNA containing the CpG islands upstream of Vis and Colos would hybridize, depending on the degree of methylation. This novel methylation chip and accompanying protocol will be available for purchase from Affymetrix in April of this year. Signals were detected using the Aerius Automated Chip Imaging System and analyzed using APT (version 1.14.3).
RESULTS
IDENTIFICATION OF VIS AND COLOS
WTSS of Hulk-1 (active Hulk) revealed exceedingly high expression of two previously non-described genes, neither of which was present in the non-active or human control lines (LCL-49, NAH-20, or NAP-9). One of these two genes (751 bp) was found to have high homology with a new gene identified in PE-1 (active Popeye), so we have presumed them to be orthologs of the same gene. This gene, henceforth called Vis, is likely responsible for the phenotypic super-human strength exhibited by both the Hulk and Popeye. Relative differences in strength between the two cases may be accounted for by polymorphisms within the sequence. The second new gene found in Hulk-1 (802 bp) was shown to have high homology with a gene identified in PS-1 (Papa Smurf). These were also presumed to be orthologs of a gene, henceforth called Colos, which is likely responsible for the intense hyper-pigmentation of both the Hulk and Papa Smurf. Again, polymorphisms may account for variation in their exact coloring.
WGSS of the Hulk genome resulted in the de novo assembly of a novel chromosome on which both Vis and Colos reside. The two genes were found to be singly exonic and on opposite arms of the chromosome.
IDENTIFICATION OF CHROMOSOME γ
Karyotypic analysis of Hulk-1 (Fig. 2a,b) revealed a collection of fragmented chromosomes (an artifact of the Hulk’s exposure to γ radiation), along with the presence of an entirely new chromosome, which we have named chromosome-γ. Both Vis and Colos localized to the γ-chromosome by FISH analysis. Contradictorily, in both PE-1 (Fig. 2c) and PS-1 (Fig. 2d), Vis and Colos localized to the X-chromosome.

Figure 2: FISH-Karyotypic analysis of the X-, γ- and hybrid chromosomes in the Hulk, Popeye and Papa Smurf. Hulk X-chromosome (a), Hulk γ-chromosome (b), Popeye X/γ-chromosome (c), and Papa Smurf X/γ-chromosome (d). Red signals depict Vis and green signals depict Colos. Chromosomes are derived from M-phase cells.
γ-irradiation and subsequent karyotyping of non-Hulk cell lines revealed the de-localization of Vis and Colos FISH signals from the X-chromosome to a now-independent chromosome-γ. We hypothesize that in normal tissues, the γ-chromosome is somehow intertwined with the centromeric region of the X-chromosome. This physical intertwining may account for previous failure to detect and sequence this chromosome. We further hypothesize that the γ-irradiation-induced dissociation of γ- from X- may arise from breakages within the telomeric region of the γ-chromosome. The discovery of this intertwining capability of chromosomes may have implications for further genomic discovery of small chromosomes associated with centromeric regions of larger chromosomes.
EPIGENETIC ANALYSIS
Hulk-1 maintained low levels of methylation in CpG islands upstream of both Vis and Colos before and after treatment with either of the two activating substances (Fig. 3a,b). Similarly, PE-1 retained low methylation upstream of the Vis gene only with and without treatment with spinach (Fig. 3a), and PS-1 had low methylation upstream of the Colos gene with and without treatment with smurf berries (Fig. 3b).
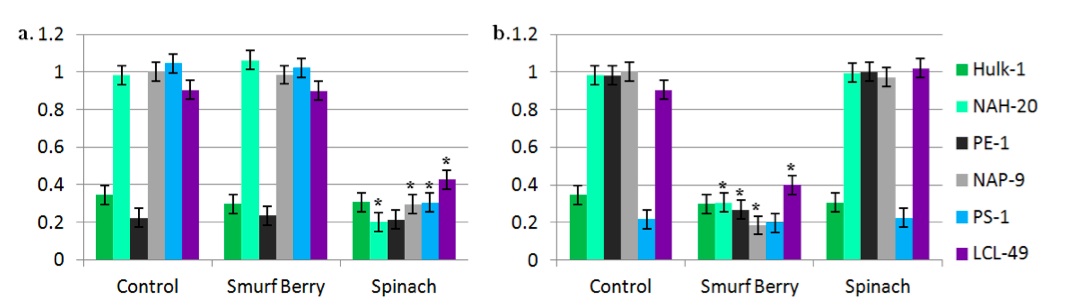
Figure 3: Methylation of CpG islands upstream of “super” genes. Normalized methylation levels of CpG islands upstream of Vis gene for strength (a) and Colos gene for color (b) following treatment with smurf berry or spinach concentrate, or untreated media. Results are shown for active and inactive Hulk, active and inactive Popeye, Papa Smurf, and human control.
In cells other than Hulk-1 and PE-1, the methylation state of CpG islands upstream of the Vis gene decreased significantly following treatment with high-dose spinach extract (Fig. 3a). Likewise, the methylation state of CpG islands upstream of the Colos gene dropped significantly following treatment of cells (other than Hulk-1 and PS-1) with low-dose smurf berry extract (Fig. 3b). Since decreased methylation of upstream CpG islands is linked to increased expression of genes [18,19], we speculate that both of these activators induce expression of their respective “super” gene by the action of a DNA methyltransferase. Further study is required to determine the sources and activation of these methyltransferases.
We can provide some suggestions for the discrepancies in modes of stimulation for our index superhero and semi-superhero cases. Smurfs live in seclusion [20] and have restricted, exclusive access to smurf berries [21] so it follows that their effects on the Colos gene are only seen within this closed population; however, spinach is a widely available commodity that is often consumed as part of a regular diet. We hypothesize that the effects of spinach on Vis are only stimulated at extremely high concentrations, well-beyond the lethal dose of iron for humans.
Popeye must bear some genetic aberration that supports a mechanism for survival of such a high dose of iron, such as a transport enzyme or metal chelator. Since the Hulk does not regularly consume either of the activator substances, we suggest that though his exposure to γ radiation has caused dissociation of the γ-chromosome from the centromere of the X- chromosome, it is only partial. The two must be further separated by some physical force, such as increased blood pressure, for the γ-chromosome to become fully active.
FUTURE DIRECTIONS
In order to understand the mechanism by which activator substances function to initiate expression of “super” genes, a full protein interactions model must be constructed. The elucidation of this pathway may promote the discovery of additional centromere-associated, novel chromosomes. This unexplored mechanism of gene regulation could be pivotal in our understanding of superhuman powers and their molecular backgrounds.
Findings from this study may be employed for curative purposes to Dr. Bruce Banner, such as si-RNA treatment or use of small inhibitors against downstream proteins.
No potential conflict of interest relevant to this article is reported.
Disclosure forms provided by the authors are available with the full text of this article at NEJM.org.
We thank Nobel Prize Laureate Dr. Dave Ng for his support.
REFERENCES
1. Lee S. The history and future of superhuman abilities. Annu Sup Rev 1973;3:33-45.
2. Siegel J and Schuster J. Genomic mutations may be behind superhe-roic abilities. DC Rev 1960;22:63-9.
3. Lee S. Unusual abilities resulting from radiation induced mutations. Sup Biol 1977;33:13-20.
4. Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet 2003;33:245-54.
5. Schafer A, Karaulanov E, Stapf U, Döderlein G, and Niehrs C. Ing1 functions in DNA demethylation by directing Gadd45a to H3K4me3. Genes and Dev 2013;27:261-73.
6. Larochelle S. Directing DNA meth-ylation. Nat Struc and Mol Biol 2013;20:308.
7. Jones P, Takai D. The role of DNA methylation in mammalian epige-netics. Science 2001;293(5332):1068-70.
8. Niehrs C. Active DNA demethylation and DNA repair. Differentiation 2009;77:1–11.
9. Schär P and Fritsch O. DNA repair and the control of DNA methylation. Prog Drug Res 2011;67:51-68.
10. Lee S. Characterizing the Hulk. Sup Biol 1974;30:21-6.
11. Lee S. Investigating the transform-ation of the Hulk. Marv Biol 1970;8:31-9.
12. Shah, S, et al. Mutational evolution in a lobular beast tumour profiled at single nucleotide resolution. Nature 2009;461:809-13.
13. Morin, R, et al. Somatic mutations altering EZH2 (Tyr641) in follicular and diffuse large B-cell lymphomas of germinal-center origin. Nat Gen 2010; 42:181-5.
14. Li, R, et al. De novo assembly of human genomes with massively parallel short read sequencing. Genome Res 2010;20:265-72.
15. Osoegawa K, Woon PY, Zhao B, Frengen E, Tateno M, Catanese JJ, and de Jong PJ. An improved approach for construction of bacterial artificial chromosome libraries. Genomics. 1998;52(1):1-8.
16. FISH protocol for Paraffin Em-bedded Tissue. Vancouver, BC.: Genetic Pathology Education Centre, 2009. (Accessed March 21, 2013, link)
17. Getoff, N, et al. Formation of sex hormone transients resulting from attack of free radicals. Anticancer Res 2013;33:941-7.
18. Razin A. CpG methylation, chromatin structure, and gene silencing — a three-way connection. EMBO J 1998;17;4905–4908.
19. Cameron E, Bachman K, Myöhänen S, Herman J, and Baylin S. Synergy of demethylation and histone deacetylase inhibition in the re-expression of genes silenced in cancer. Nat Gen 1999;21:103–107.
20. Banfly A, VanDeely F, Shaw S, and Culliford P. Observations of Smurfs in their Natural Habitat. Spirou Mag 1959;5:32-37.
21. Tressens D, Sagely K, Staffords J, and Culliford. Exploration of the Smurf Diet. Spirou Mag 1962;1:12-23.
