RIBOSWITCHES: REFORMING OUR UNDERSTANDING OF METABOLIC REGULATION
The biochemical potential of a cell to carry out specific chemical reactions is nothing short of enormous. Even the simplest of cells have the ability to catalyze over a thousand reactions, only a subset of which is required at any given time.
In order to save energy and resources, the cell needs to regulate these reactions such that only those that are necessary are carried out. In simplest terms, this involves a two-component system: one for sensing environmental conditions and thus metabolic needs, the other for translating these needs into a regulatory mechanism that can induce or suppress pathways according to the requirements of the cell.
Until very recently, these two functions were thought to be carried out exclusively by proteins. One common prototype for protein-mediated regula-tion is the E. coli trp operon, which regulates the cell’s ability to synthesize the amino acid tryptophan when it is not available from the environment. It involves direct binding of a repressor protein, TrpR, to DNA at the trp promoter site, physically occluding RNA polymerase from initiating transcription at that site (Fig 1a).
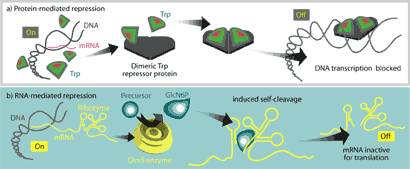
Figure 1. – Comparison of protein-mediated metabolic regulation and the riboswitch mechanism. a. In the trp operon, DNA is available for transcription of mRNA when only low levels of tryptophan are available. Presence of tryptophan from the environment causes a conformational change in the repressor protein that allows it to bind DNA at the promoter and repress transcription of the operon. b. The GlmS protein is an important enzyme of the glucosamine-6-phosphate (GlcN6P) synthesis pathway. A regulatory region of mRNA is found downstream of the coding region for GlmS. When no GlcN6P is present in the medium, the mRNA is translated and the GlmS synthesizes GlcN6P. Higher levels of GlcN6P lead to increased binding of the product to the regulatory region. The conformational change caused by binding induces auto-cleavage of the glmS mRNA, rendering it non-functional. Although the coding region for the protein is not affected, loss of the upstream region seems to prevent initiation of translation (From 7).
Recently, however, the unchallenged supremacy of proteins in metabolic regulation has been called into question through the discovery of riboswitches. These specialized stretches of non-coding mRNA can specifically bind their cognate metabolites, inducing conformational changes and/or ribozyme activity that lead to physical occlusion of the ribosome, transcription attenuation, or self-cleavage of the mRNA (Fig 1b). In this way, mRNAs can directly regulate the biosynthesis of protein products associated with the metabolites that bind them.
Suspiciously Elegant
The discovery of riboswitches was nearly as exceptional as the mechanism itself. In 1999, Ron Breaker of Yale University and his team were working on a class of synthetic RNAs, called aptamers, which bind to small molecules. It occurred to Breaker that the genetic methods that his group was exploiting to create these biosensors were so elegant and effective that it was surprising they hadn’t been observed in vivo (14). This speculation led to a search for natural riboswitch mechanisms.
The first natural riboswitches were found in genes involved in vitamin biosynthesis. These pathways had been somewhat perplexing to biologists, since no regulatory proteins had yet been associated with them (39). Investigations by Winkler et al and Nahvi et al found riboswitch regulation in B1 (46) and B12 (26) biosynthesis, respectively.
Intriguingly, Harold White III had suggested twenty-five years earlier that this type of binding between nucleic acids and coenzymes could be an artefact of a pre-protein world (44).
Relics of the Past?
Indeed, the discovery of riboswitches has been an exciting and confirming revelation for those biologists who believe early life was based on an RNA genome. This world is thought to have pre-dated DNA and protein and may have existed as many as four billion years ago (12).
The functional diversity of RNA has been appreciated for years; scientists have known of its roles as a carrier of genetic information and nucleic acid, amino acid adapter in translation, a primer for DNA replication, and a structural component in the ribosome. This ability of RNA to carry out so many essential cell functions, and particularly its pervasiveness in protein synthesis, is suggestive that current biological systems have their origins in an ‘RNA world.’ In addition to the afore-mentioned repertoire, catalytic activity was attributed to RNA in the early 1980’s with the discovery of ribozymes (16). Thus, it is plausible that RNA could have supported many of the essential roles presently represented by DNA and protein.
Although the discovery of ribozymes bolstered the theory of an ancient biological system based on RNA, some critical questions remained unanswered. Among them was how gene expression had been regulated according to signals from the environment if proteins had not yet evolved. Riboswitches seem to have answered this question.
The presence of riboswitches across the domains further supports the theory that they were involved in early forms of gene regulation. Although at this point, most known riboswitches have been identified in bacteria, there is emerging evidence of these systems in archaea and eukaryotes as well, particularly in the case of the THI-box riboswitch (17, 37, 43). Known riboswitches and their hosts are detailed in Table 1. Note that many of the organisms known to contain riboswitches are those that are well- studied (Bacillus) and have been subject to searches. Essentially, riboswitches can only be found where they are sought, and there is great promise that the host range for this unique genetic control system will expand with the appreciation of its significance in the scientific community.
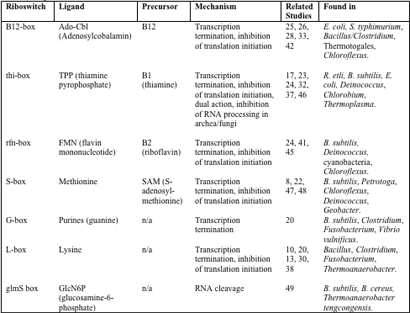
Table 1 – Overview of known riboswitches and their mechanisms (integrated from 18, 29, 37, 40, 43 and 49)
Methods of Action
Despite the as yet still limited study of natural riboswitches, these mRNAs have proven themselves to be quite variable in their methods of action. Known methods include formation of stem-loops that lead to transcription attenuation (occlusion of RNA polymerase), inhibition of translation initiation (exclusion of ribosome at Shine-Dalgarno site, or RBS), or, as more recently seen in the glmS riboswitch, cleavage of mRNA (49).
It appears that organisms use different mechanisms in the functioning of the same riboswitch (35). As in traditional protein regulation, the trend seems to be that Gram-positive bacteria tend towards transcriptional regulation and Gram-negatives towards translational regulation (4, 41). However, this is merely a trend and not a rule, and the regulatory mechanisms may be seen in any bacterium, regardless of Gram stain characteristics. Indeed, two mechanisms may even act to regulate the same riboswitch element in the same cell. This appears to be the case in the B. subtilis RFN element, in which binding of flavin mononucleotide (FMN) acts on the rib operon mRNA via transcription termination but acts on the ypaA transcript via sequestration of the RBS (45).
In all cases, specific, high affinity binding of a metabolite effector to a highly conserved non-coding region of its cognate mRNA initializes a conformational change in the mRNA that leads to the formation of a stem-loop structure. Where this stem-loop forms, the characteristics of the region involved determine the overall effect of the binding. Figure 2 illustrates the known possibilities, which will be discussed below.
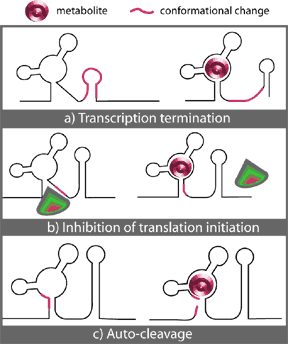
Figure 2. – Three known mechanisms of riboswitch action upon binding of metabolite (M): a) Transcription termination. b) Inhibition of translation initiation. c) Auto-cleavage. (From 35.)
Transcription termination
In conditions of low effector concentrations, these transcripts have an anti-terminator in their secondary structure that prevents the formation of a terminator loop. Binding of a metabolite to a nearby riboswitch element in the mRNA induces a change in secondary structure that destabilizes the hairpin anti-terminator in the mRNA. As the anti-terminator dissociates, the sequence formerly part of the stem is revealed and allowed to pair with a terminator sequence (Fig. 2a). The resultant terminator loop is usually located in the 5’ UTR and is often followed by a downstream polyuridine tract (18). The terminator loop thus reduces the stability of either the mRNA:RNA polymerase interaction and/or of the DNA:RNA hybrid in a rho-independent manner (27). This causes the RNA polymerase to dissociate, terminating transcription prematurely.
Inhibition of Translation Initiation
Alternatively, riboswitches may act at the level of translational control through the sequestration of the ribosome binding sequence in the mRNA. When no metabolite is bound, the Shine-Dalgarno (SD) site is exposed and the ribosome can bind and initiate translation. Binding of the metabolite to the 5’ leader region of the mRNA induces the formation of an SD:anti-SD stem-loop structure that masks the ribosome binding site such that initial step of translation, binding of the ribosome to the mRNA, is not achieved (Fig. 2b).
Auto-cleavage
Auto-cleavage is the most recently-discovered and possibly the most remarkable riboswitch mechanism known thus far. Whereas the previous two mechanisms have each been observed in multiple riboswitches and in a variety of bacterial systems, the only riboswitch with ribozyme action known to date is the glmS-box discovered by Winkler and his group as recently as 2004 (49).
Located in the 5’ untranslated region of the glmS gene, this riboswitch is astonishingly specific to its ligand, glucosamine-6-phosphte, whose binding increases the rate of cleavage 1,000 fold (49). The precise mechanism of cleavage remains unknown, but Winkler has proposed that it is accomplished through internal phosphoester transfer, which he noted has been studied in other known riboswitches (49). In short, it seems that the conformational change induced by the binding of the ligand to the riboswitch brings adjacent nucleotides in line with each other in an orientation that favours cleavage (Fig. 2c).
Other Methods of Action
All of the riboswitch effects described above have related to repression of gene function upon binding of a specific metabolite involved somehow in that gene’s function. It has, however, also been suggested that the same mechanisms could be involved in positive gene regulation as well (18, 29). In these cases, binding of a metabolite would release the terminator hairpin or liberate the SD-site, permitting full transcription or translation, respectively. Although this type of control has not yet been observed in natural systems, there is no reason to rule it out.
There is also speculation that eukaryotic riboswitches have the potential for more complex methods of regulation. For example, it has been suggested the riboswitches could also potentially play a role in the processing and transport of mRNA in eukaryotes (29). Again, this has not yet been observed, but bearing in mind how recent is the discovery of riboswitches, it may just be a matter of time. Our understanding of these elegant control systems is only just beginning to develop.
Identifying a Riboswitch
Several techniques are available to geneticists to detect, confirm, and investigate the presence of new riboswitches. Some of the most popular experiments include sequence analysis, lacZ fusions, and in-line probing and equilibrium dialysis, respectively, and will be discussed below.
Sequence analysis
As mentioned above, known riboswitch elements are well-conserved across genera (9). This makes sequence analysis a particularly useful tool in searching for new riboswitches. At present, at least two databases are available to investigate the sequences of putative riboswitches: Breaker Lab Intergenic Sequence Server (BLISS) and the Riboswitch Finder.
These databases are based on known riboswitch sequences and scan entries for related motifs. They are also able to predict the structure of the putative riboswitches, can evaluate the statistical probability of a positive identification, and have a low false-positive rate (3). The success of these search engines has been attributed to the pervasiveness of riboswitches throughout prokaryotic regulation of metabolism (3).
LacZ fusions
Once an RNA sequence has been singled out as a candidate for a new riboswitch, tests must still be carried out to substantiate the hypothesis. A simple and popular test is the lacZ fusion (20, 46).
The upstream region of the gene of interest is amplified by PCR. The product, which contains the proposed riboswitch region, is ligated into a plasmid upstream of and in-frame with the lacZ gene. ß-galactosidase expression should now be under control of the putative riboswitch.
Cells are transformed with the vector and successful transformants are found using a selectable marker present on the plasmid (for example, ampicillin resistance). Inducers can then be added that are known to be involved in the regulation of the original gene, and the effect can be measured with ß-galactosidase enzyme assays to detect the levels of LacZ in the cell. Sequencing should also be performed to confirm the identity of the regulatory region – this is done to test the integrity of the ligation reaction.
This type of assay is particularly useful in determining the specificity of a riboswitch for its ligand by using various analogs and comparing their effects. Many riboswitches are so specific that they will distinguish between something as small as an amino group exchanged for a hydroxyl group (49).
In-line Probing
This is a method to more accurately identify the 5’ and 3’ ends of the riboswitch region and calculate the size and apparent dissociation constant, KD, of the ligand for its binding site. The KD of riboswitches is completely analogous to the KD of protein regulation systems, and represents the concentration of ligand that leads to a state of half-saturation of the binding site.
In-line probing relies on the fact that there is a natural rate of spontaneous cleavage within RNA. Cleavage occurs when a phosphodiester linkage is subjected to internal nucleophilic attack by the 2’ oxygen adjacent to and in-line with it (34). Structured regions of RNA, such as those in the base-paired stems of riboswitch stem-loops, are less susceptible to spontaneous cleavage than non-structured regions (25).
Therefore, assessing the cleavage products of a given riboswitch sequence in the absence of its cognate ligand can yield helpful information about the size, structure and precise location of the riboswitch. Such analyses are accomplished by amplifying the riboswitch region by PCR, making RNA copies of the PCR product, subjecting the products to cleavage, separating the resultant fragments with polyacrylamide gel electrophoresis (PAGE), and sequencing them (20, 25). An extension of this technique involves calculating the relative amounts of cleavage products in varying concentrations of metabolite ligand to calculate the KD of the ligand:riboswitch complex (25, 34).
Equilibrium Dialysis
This is another popular technique used to identify which ligand binds a particular riboswitch, and what the KD for that ligand is. The experimental setup consists of two chambers separated by a membrane: One chamber contains a solution of the riboswitch element, the other contains a radiolabelled form of the metabolite of interest (20). If the metabolite has binding affinity for the riboswitch, its concentration will shift towards the chamber with the riboswitch solution. If not, there will be no change in concentration. Higher binding affinities attract higher concentrations of metabolite towards the chamber with the riboswitch solution.
This, along with the lacZ fusion method, is a simple and convenient way to test the specificity of a riboswitch for its ligand.
Putting it all together: the RFN-box
The riboflavin (RFN) riboswitch was one of the first to be discovered and also appears to be one of the most common in natural systems (21). It serves as a good example of the steps involved in discovering and investigating a new riboswitch, as well as the mechanism of action in vivo.
At the beginning of the 1990’s, riboflavin, or vitamin B2, was part of the perplexing puzzle of vitamin biosynthesis regulation. Although it was clear that negative regulation of genes involved in synthesis of vitamins was occurring, there was no “smoking gun”(18); no repressor proteins had yet been identified for these pathways.
However, what had been recognized was that the 5’ UTR of the rib operon was well conserved throughout Gram positive bacteria (9), and that mutations in this region led to the loss of negative control and over-production of riboflavin in the cell (11, 15). It was also known to have extensive secondary structure, folding into five hairpins with well-conserved sequences in the base pairs of the stems (9) (Fig. 3).
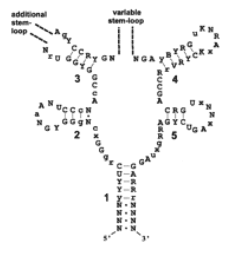
Figure 3 – The structure of the RFN element showing invariant bases (capitalized), well-conserved bases (lower case), and variable bases (R, Y, K, B, V, N or X) (From 41)
Taking a cue from studies of synthetic aptamers that bound RFN in vitro (1, 19, 31), Mikhail Gelfand was the first to suggest that this region may directly bind a metabolite regulator.
Shortly thereafter, Vitreschak et al published their results of an extensive sequence comparison study done on RFN elements (41). They found that these elements were present both in Gram positive and Gram negative bacteria, but their location and the arrangement of the rfn genes were somewhat different in each case.
In Gram positive organisms such as Bacillus, the rfn genes were arranged in operons, which contained genes coding for both metabolic and transport-related proteins. In most cases, each operon was preceded by an RFN element followed by a run of thymidines, indicating a potential transcription terminator.
By contrast, Gram negatives tended to have rfn genes distributed singly throughout the genome. These were more often preceded by an RFN element that overlapped the RBS, suggesting the presence of a sequestor for inhibition of translation initiation.
Though this study provided more insight into the pervasiveness of RFN elements across genera, as well as the regulation of rfn genes, it did nothing to confirm or deny the direct binding of metabolites to the RFN box. This was confirmed a few months later in a study by Winkler’s group (45).
This study took advantage of in-line probing to show that the structure of the RFN element was altered by addition of flavin mononucleotide (FMN, a riboflavin analog and derivative) to a solution in the absence of protein. Furthermore, the dissociation constants for FMN, riboflavin, and flavin adenine dinucleotide (FAD) were measured and were found to be quite different, at KD = 5 nM, 3 µM, and 300 nM respectively. Amazingly, the phosphate group that differentiates FMN from riboflavin seems to make a 1,000 fold difference to the binding affinity of the ligand for the RFN element. This element was clearly very specific for its ligand.
Sequence analyses revealed yet another interesting twist to this element. While the majority of the rfn genes in B. subtilis were under riboswitch-mediated transcriptional control on the rfn operon, a lone gene, ypaA, believed to code for a transporter involved in riboflavin biosynthesis, was under control of a different riboswitch. The location of this riboswitch indicated a role in the sequestration of the RBS, that is, in translational control. This may be evidence of horizontal gene transfer of riboswitches between organisms.
Thus, Winkler’s study was the first to conclusively show that genes for riboflavin biosynthesis and transport were under control of direct metabolite-binding to mRNA, and that the region where this occurred corresponded to the well-conserved 5’ leader sequence known as the rfn element. This group’s findings were confirmed later in the same year by a similar study conducted by Alexander Mironov (24).
Implications and Applications
Since the discovery of riboswitches only a few years ago, at least seven different riboswitches have been found to exist in dozens of organisms spanning the kingdoms. It has been estimated that at least 68 genes in B. subtilis (2% of the genome) are under the control of these mRNA regulatory sequences (36). When one considers the elegance in the simplicity of the system, the potential for rapid response times, and the economics of not having to make a protein, it may be surprising that these elements are not more widespread. But the fact is that proteins did evolve, and thus there must have been a motivation for them. One advantage of proteins is that they provide more variation and flexibility than their polynucleotide cousins: there are 20 amino acids compared to just four ribonucleotides. Furthermore, their ability to act in trans allows for the biological cross-talk and intricate signalling pathways that are so important to regulatory networks. Clearly, riboswitches do not contain all the answers, but they have been overlooked for years and are now receiving the recognition they deserve.
What kind of future lies ahead for these unique mRNAs? Antibiotic potential has been suggested as one possibility (6, 48). Given the central role of riboswitches in essential cell metabolism, it is conceivable that they could be exploited to the detriment of their host. Synthetic analogs of riboswitch ligands could be engineered to shut off central metabolic pathways, arresting the growth of the bacteria. There is a hope that this type of antimicrobial treatment would be less toxic than the alternatives, since RNA is targeted instead of protein (18).
They could also be used for the same purposes as the synthetic aptamers that led to their discovery: as molecular chemosensors for measuring chemical composition or biochemical secretions (5). Given that the affinity for ligand in the natural systems is so much higher than their synthetic counterparts (29), this could develop into a sensitive new branch of biotechnology with applications in research and medicine, among other fields.
A further possibility is that of using riboswitch fusions to trans-genes as a means to regulate gene inserts through small molecule inducers (18). This could have widespread applications in genetic research, and even in medicine and gene therapy.
Yet another possible application is the use of riboswitches in taxonomic studies. Though regions of riboswitches are well-conserved, there are distinct variable regions that have been indicated as being dependant on taxonomy (41).
A better understanding of this relationship could lead to the use of riboswitches as another tool in determining the evolutionary relationships between organisms.
Probably the biggest task in current riboswitch research is getting a grip on just how widespread this mechanism is. If these elements are indeed relics of an ancient RNA world, then they have been under our noses all along, waiting to be discovered. It is humbling to think that RNA has surprised us yet again in its spectrum of capabilities. It leads one to wonder how much we have missed. There is a sense that the seven known riboswitches are just the tip of the iceberg.
It is difficult to say what time and research will reveal about the scope of riboswitch control in prokaryotes and eukaryotes alike, but one thing is certain – riboswitches have finally earned their spot in the repertoire of metabolic regulation mechanisms.
References
1. Araki, M., Y. Okuno, Y. Hara, and Y. Sugiura. 1998. Allosteric regulation of a ribozyme activity through ligand-induced conformational change. Nucleic Acids Res. 26:3379-3384.
2. Barrick, J. E., K. A. Corbino, W. C. Winkler, A. Nahvi, M. Mandal, J. Collins, M. Lee, A. Roth, N. Sudarsan, I. Jona, J. K. Wickiser, and R. R. Breaker. 2004. New RNA motifs suggest an expanded scope for Riboswitches in bacterial genetic control. Proc. Natl. Acad. Sci. 101:6421-6426.
3. Bengert, P., and T. Dandekar. 2004. Riboswitch finder – a tool for identification of riboswitch RNAs. Nucleic Acids Res. 32:154-159.
4. Brantl, S. 2004. Bacterial gene regulation: from transcription attenuation to riboswitches and ribozymes. Trends Microbiol. 12:473-475.
5. Breaker, R.R. 2002. Engineered allosteric ribozymes as biosensor components. Curr. Opin. Biotechnol. 13:31-39.
6. Breaker, R. R., S. Cohen-Chalamish, G. M. Emilsson, S. Nakamura, N. Sudarsan, and W. C. Winkler. 2003. Riboswitch RNAs: A new class of antimicrobial targets. [Meeting] Abstracts of the Interscience Conference on Antimicrobial Agents & Chemotherapy. 43:94-95.
7. Cech, T.R. 2004. RNA finds a simpler way. Nature. 428:263-264.
8. Epshtein, V., A. S. Mironov, and E. Nudler. 2003. The riboswitch-mediated control of sulfur metabolism in bacteria. Proc. Natl. Acad. Sci. U.S.A. 100:5052-5056.
9. Gelfand, M. S., A. A. Mironov, J. Jomantas, Y. I. Kozlov, and D. A. Perumov. 1999. A conserved RNA structure element involved in the regulation of bacterial riboflavin synthesis. Trends Genet. 15:439-442.
10. Grundy, F. J., S. C. Lehman, and T. M. Henkin. 2003. The L box regulon: Lysine sensing by leader RNAs of bacterial lysine biosynthesis genes. Proc. Natl. Acad. Sci. U.S.A. 100:12057-12062.
11. Gusarov, I. I., R. A. Kreneva, K. V. Rybak, D. A. Podcherniaev, I. uV. Iomantas, L. G. Kolibaba, B. M. Polanuer, I. uI. Kozlov, and D. A. Perumov. 1997. Primary structure and functional activity of the Bacillus subtilis ribC gene. Mol. Biol. 31:446-453.
12. Joyce, G. F. 2002. The antiquity of RNA-based evolution. Nature. 418:214-221.
13. Kochhar, S. and H. Paulus. 1996. Lysine-induced premature transcription termination in the lysC operon of Bacillus subtilis. FEMS Microbiol. Lett. 71:23-27.
14. Knight, J. 2003. Switched on to RNA. Nature. 425:232-233.
15. Kreneva, R. A. and D. A. Perumov. 1990. Genetic mapping of regulatory mutations of Bacillus subtilis riboflavin operon. Mol. Gen. Genet. 222:467-469.
16. Kruger, K., P. J. Grabowski, A. J. Zaug, J. Sands, D. E. Gottschling, and T. R. Cech. 1982. Self-splicing RNA: Autoexcision and autocyclization of the ribosomal RNA intervening sequence of Tetrahymena. Cell. 31:147-157.
17. Kubodera, T., M. Watanabe, K. Yoshiuchi, N. Yamashita, A. Nishamura, S. Nakai, K. Gomi, and H. Hanamoto. 2003. Thiamine-regulated gene expression of Aspergillus oryzae thiA requires splicing of the intron containing a riboswitch-like domain in the 5’-UTR. FEBS Lett. 555:516-520.
18. Lai, E.C. 2003. RNA Sensors and Riboswitches: Self-Regulating Messages. Curr. Biol. 13:R285-291.
19. Lauhon, C. T. and J. W. Szostak. 1995. RNA aptamers that bind flavin and nicotinamide redox cofactors. J. Am. Chem. Soc. 117:1246-1257.
20. Mandal, M. B. Boese, J. E. Barrick, W. C. Winkler, and R. R. Breaker. 2003. Riboswitches Control Fundamental Biochemical Pathways in Bacillus subtilis and Other Bacteria. Cell. 113:577-586.
21. Mandal, M. and R. R. Breaker. 2004. Gene Regulation by Riboswitches. Nat. Rev. Mol. Cell Biol. 5:451-463.
22. McDaniel, B. A. M., F. J. Grundy, I. Artsimovitch, and T. M. Henkin. 2003. Transcription termination control of the S box system: Direct measurement of S-adenosylmethionine by the leader RNA. Proc. Natl. Acad. Sci. U.S.A. 100:3083-3088.
23. Miranda-Rios, J., M. Navarro, and M. Soberón. 2001. A conserved RNA structure (thi box) is involved in regulation of thiamine biosynthetic gene expression in bacteria. Proc. Natl. Acad. Sci. U.S.A. 98:9736-9741.
24. Mironov, A. S., I. Gusarov, R. Rafikov, L. E. Lopez, K. Shatalin, R. A. Kreneva, D. A. Perumov, and E. Nudler. 2002. Sensing small molecules by nascent RNA: a mechanism to control transcription in bacteria. Cell. 111:747-756.
25. Nahvi, A., N. Sudarsan, M. S. Ebert, X. Zou, K. L. Brown, and R. R. Breaker. 2002. Genetic control by a metabolite binding mRNA. Chem. Biol. 9:1043-1049.
26. Nahvi, A., J. E. Barrick, and R. R. Breaker. 2004. Coenzyme B12 riboswitches are widespread genetic control elements in prokaryotes. Nucleic Acids Res. 32:143-150.
27. Nelson, D. L. and M. M. Cox. 2000. Lehninger Principles of Biochemistry. 3rd ed. New York: Worth.
28. Nou, X. and R. J. Kadner. 2000. Adenosylcobamalin inhibits ribosome binding to btuB RNA. Proc. Natl. Acad. Sci. U.S.A. 97:7190-7195.
29. Nudler, E., and A. S. Mironov. 2004. The riboswitch control of bacterial metabolism. Trends Biochem. Sci. 29:11-17.
30. Patte, J. C., M. Akrim, and V. Mejean. 1998. The leader sequence of the Escherichia coli lysC gene is involved in the regulation of LysC synthesis. FEMS Microbiol. Lett. 169:165-170.
31. Patel, D. J., A. K. Suri, F. Jiang, L. Jiang, P. Fan, R. A. Kumar, and S. Nonin. 1997. Structure, recognition, and adaptive binding in RNA aptamers complexes. J. Mol. Biol. 272:645-664.
32. Rodionov, D.A. Vitreschak, A. G., A. A. Mironov, and M. S. Gelfand. 2002. Comparative genomics of thiamine biosynthesis in procaryotes. New genes and regulatory mechanisms. J. Biol. Chem. 277:48949-48959.
33. Rodionov, D.A. Vitreschak, A. G., A. A. Mironov, and M. S. Gelfand. 2003. Comparative Genomics of the Vitamin B12 Metabolism and Regulation in Prokaryotes. J. Biol. Chem. 278:41148-41159.
34. Soukup, G. A. and R. R. Breaker. 1999. Relationship between internucleotide linkage geometry and the stability of RNA. RNA. 5:1308-1325.
35. Soukup, J. K., and G. A. Soukup. 2004. Riboswitches exert genetic control through metabolite-induced conformational change. Curr. Opin. Struct. Biol. 14:344-349.
36. Stormo, G.D. 2003. New Tricks for an Old Dogma: Riboswitches as cis-Only Regulatory Systems. Mol. Cell. 11:1419-1423.
37. Sudarsan, N., J. E. Barrick, and R. R. Breaker. 2003. Metabolite-binding RNA domains are present in the genes of eukaryotes. RNA. 9:644-647.
38. Sudarsan, N., J. K. Wickiser, S. Nakamura, M. S. Ebert, and R. R. Breaker. 2003. An mRNA structure in bacteria that controls gene expression by binding lysine. Genes Dev. 17:2688-2697.
39. Szostak, J. W. 2002. RNA gets a grip on translation. Nature. 419:890-891.
40. Templeton, G. W. and G. B. G. Moorhead. 2004. A Renaissance of Metabolite Sensing and Signalling: From Modular Domains to Riboswitches. Plant Cell. 16:2252-2257.
41. Vitreschak, A. G., D. A. Rodionov, A. A. Mironov, and M. S. Gelfand. 2002. Regulation of riboflavin biosynthesis and transport genes in bacteria by transcriptional and translational attenuation. Nucleic Acids Res. 30:3141-3151.
42. Vitreschak, A. G., D. A. Rodionov, A. A. Mironov, and M. S. Gelfand. 2003. Regulation of the vitamin B12 metabolism and transport in bacteria by a conserved RNA structural element. RNA. 9:1084-1097.
43. Vitreschak, A. G., D. A. Rodionov, A. A. Mironov, and M. S. Gelfand. 2004. Riboswitches: the oldest mechanism for the regulation of gene expression? Trends Genet. 20:44-50.
44. White, H. B. III. 1976. Coenzymes as Fossils of an Earlier Metabolic State. J. Mol. Evol. 7:101-104
45. Winkler, W., S. Cohen-Chalamish, and R. R. Breaker. 2002. An mRNA structure that controls gene expression by binding FMN. Proc. Natl. Acad. Sci. U. S. A. 99:15908-15913.
46. Winkler, W., A. Nahvi, and R.R. Breaker. 2002. Thiamine derivatives bind messenger RNAs to regulate bacterial gene expression. Nature. 419:952-956.
47. Winkler, W., A. Nahvi, N. Sudarsan, J. E. Barrick, and R. R. Breaker. 2003. An mRNA structure that controls gene expression by binding S-adenosylmethionine. Nat. Struct. Biol. 10:701-707.
48. Winkler, W. C., A. Nahvi, S. Nakamura, N. Sudarsan, J. E. Barrick, and R. R. Breaker. 2003. Bacterial mRNAs that bind S-adenosylmethionine as targets for drug discovery. [Meeting] Abstracts of the Interscience Conference on Antimicrobial Agents & Chemotherapy. 43:95
49. Winkler, W., A. Nahvi, A. Roth, J. A. Collins, and R. R. Breaker. 2004. Control of gene expression by a natural metabolite-responsive ribozyme. Nature. 428:281-286.