THE LAST GREAT UNCONTROLLED PLAGUE OF MANKIND
Introduction:
Influenza remains an important disease in humans and animals. In contrast to measles, smallpox and poliomyelitis, influenza is caused by viruses that undergo continuous antigenic change and that possess an animal reservoir. Thus, new epidemics and pandemics are likely to occur in the future, and eradication of the disease will be difficult to achieve. Although it is not clear whether a new pandemic is imminent, it would be prudent to take into account the lessons we have learned from studying different human and animal influenza viruses. Influenza has long been with us; indeed, the name itself refers to the ancient belief that it was caused by a malign and super natural influence. In Florence during the time of the Renaissance, astrologers linked a curious juxtaposition of stars with an outbreak of infection in the city and attributed it to the “influence” of the stars, hence influenza. Known in the sixteenth century as “the newe Acquayntance”, influenza still causes major outbreaks of acute respiratory infection.
Classification:
Although laymen refer to many incapacitating respiratory infection as “flu”, true influenza is caused by the small family of the Orthomyxoviridae. Myxo derives from the Greek for mucus and refers to the ability of these viruses to attach to mucoproteins on the cell surface; ortho means true or regular, as in orthodox, and distinguishes these viruses from the Paramyxoviridae (measles is a member of this family). There are four genera; distinguished serologically on the basis of their matrix (M) and nucleoprotein (N) antigens. They are: Influenza virus A, B and C as well as Thogoto-like virus which is a tick-borne virus of mammals.
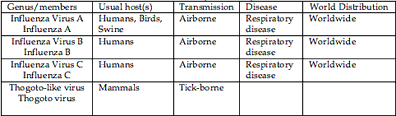
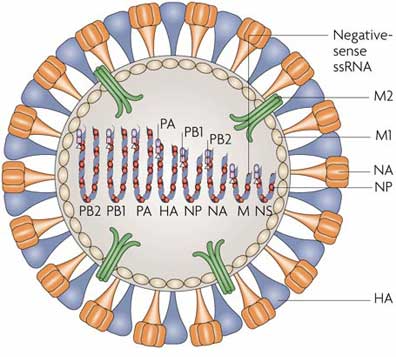
Influenza viruses A and B are closely related, but influenza A infects a wide spectrum of birds and mammals including humans, whereas influenza B infects only humans. Influenza C is more divergent. Eight segments of RNA, totaling about 14 kb, comprise the genomes of influenza A and B viruses whereas influenza C has only 7 segments. All three influenza viruses infect man and cause disease, but influenza A represents the most serious human pathogen because it causes very large, recurrent epidemic and even pandemic with significant mortality. The RNA is closely associated with the nucleoprotein (NP) to form a helical structure. There are 4 antigens present, the haemagglutinin (HA), neuraminidase (NA), nucleocapsid (NA), the matrix (M) and the nucleocapsid proteins (NP). Two surface glycoproteins are seen on the surface as rod-shaped projections. HA mediates the attachment of the virus to the cellular receptor. Influenza A viruses have been designated on the basis of the antigenic relationships of the external spike haemagglutinin (HA) and neuraminidase (NA) proteins. In earlier years HA and NA antigens driving from birds and other animals were given appropriate letters (for instance Hsw for haemagglutinin of a swine –type virus or Nav for a neuraminidase of avian origin). Since 1980 the antigens have been given simple sequential numbers, H1-H15 and N1-N9. Of these only H1, H2, or H3 and N1 or N2 are known to infect humans but do not appear to spraed from person to person. You could see following figure as World Health Organization nomenclature for influenza viruses.

Type B strains are designated on the same system, but without H and N numbers since major changes in these antigens have so far not been observed.
Morphology:
The virions are 100-200 nm in diameter and are more or less spherical. The lipid envelope is covered with about 500 projection spikes, which can be seen clearly under the electron microscope. About 80 percent of them are haemagglutinin antigen and the reminders are another antigen, neuraminidase, and have a mushroom-like shape. The haemagglutinin (HA) is a rod-shaped glycoprotein with a triangular cross-section. It was first identified by its ability to agglutinate erythrocytes, hence its name, but it is now apparent that it also has important roles in the attachment and entry of virus to the cells of the host and in determining virulence. The neuraminidase (NA) can remove neuraminic (sialic) acid from receptor proteins. Its main function seems to be connected with release of new virus from cells.
Clinical Features and pathological aspects:
There are no difference between Influenza A and B as regards the clinical picture. After an incubation period of 2-3 days there is usually a very abrupt onset with shivering, malaise, headache, and aching in the limbs and back. Characteristically, the patient is prostrated and has to take to bed. The temperature rises rapidly to around 39 C. Influenza is not characterized by runny noses or sore throats at the beginning, as are common cold infections. Fortunately, influenza is usually short-lived in younger persons. In older people and the at-risk group, however, recovery may take much longer, with persistent weakness and lassitude sometimes for 3-6 months. In general, the severity of influenza is proportional to age. Apart from secondary bacterial infection there are few complications, but one are condition, Reye’s syndrome, is sometimes associated with influenza in children, often of the B type. The taking of aspirin has also implicated in the causation of this syndrome, which involves encephalopathy with fatty degeneration of the liver and other viscera; it is often fatal.
Pathogenesis: Infection is acquired by the respiratory route and is usually an infection of the upper respiratory tract. Virus multiplies in the epithelial cells in the nose and sinus passages and destroys the cilia, which are an important element in the defense of the respiratory system. Viral infection of the lower respiratory tract, in the form of influenza pneumonia, sometimes occurs, presenting as an overwhelming toxemia with higher mortality. The virus replicates in epithelial cells of the alveoli, causing exudation into the air sacs and pneumonia. Pneumonia is, however, often due to secondary infection with bacteria.
Reservoirs of influenza A in nature:
Influenza A viruses infect a variety of animals, including humans, pigs, horses, sea mammals, and birds. Recent phylogenetic studies of influenza A viruses have revealed species-specific lineages of viral genes and have demonstrated that the prevalence of interspecies transmission depends on the animal species. They have also revealed that aquatic birds are the source of all influenza viruses in other species.
Genetic variation in influenza viruses:
Influenza A viruses readily undergo gene “swapping” or reassortment, so that, in a cell infected simultaneously with two different viruses, the progeny virions may contain mixtures of each parent’s genes. Add this property to the ability of influenza A virus to infect animals such as pigs and birds that often live in close association with humans, and we have a situation in which double infections with viruses of human and non-human origin may result at unpredictable intervals in the formation of new strains with genetic compositions differing from those in general circulation.
This reassortment of genes known as antigenic shift, can, of course, also take place between two influenza A viruses of human origin. It cannot occur between influenza A and B viruses. RNA viruse tend to have high mutation rate-more than 10.000 times higher than that of human or viral DNA- and this is true of all the influenza viruses. The viral RNA replicase is a low-friendly enzyme, so transcription errors accumulate. Moreover there are no proof-reading of corrective enzymes. These mutations give rise to changes in the viral polypeptides, such as HA which, out of a total of 250 amino acids, undergoes two or three amino-acid substitutions each year. Both influenza A and B are subject to antigenic drift but only A viruses undergo antigenic shift and hence have the potential of causing pandemics.
Epidemiology of influenza viruses:
The annual death rate in the United States from influenza A in people over 65 is 1 per 2200, and in an epidemic year the death rate may be 1 in 300. Descriptions of epidemics and pandemics of influenza have been recorded for over four centuries. The rapid, global spread of pandemic influenza may be a relatively modern development related to increases in population and the growth of transportation systems necessary for the global transmission of the novel virus. Animals may have played a crucial role in past influenza epidemics as well as in modern pandemics. Outbreaks of respiratory disease among horses were recorded concurrently with outbreaks in humans during the eighteenth and nineteenth centuries, and in recent years it has been suggested that swine and birds are prominently involved in the generation of influenza pandemics.
The Asiatic flu, 1889-1890: It was the last great pandemic of the nineteenth century. The first report came from Russia in May 1889. It rapidly spread west and hit North America in December 1889, South America in February–April 1890, India in February-March 1890, and Australia in March–April 1890. It was purportedly caused by the H2N8 type of flu virus and had a very high attack and mortality rate.
The Spanish Flu, 1918-1920 [A (H1N1)]: Although the very young and elderly are normally at the most risk from influenza, the influenza pandemic of 1918-1919 was unusual in that mortality was high in health young adults. The dramatic increase in the death rate in the 20-to 29-year-old group in 1918, in which people of this age were more likely to die than the old and the young, is striking. In 1918 children would skip rope to the rhyme: I had a little bird, Its name was Enza, I opened the window, And in-flu-enza. The epidemic caused by this extremely virulent virus spread around the world over a period of about a year and ultimately infected an estimated 20% of the world’s population. The overall mortality was perhaps 2% but in some regions of the world, for example, regions of Central America and certain islands in the Pacific, 10-20% of entire population died in the epidemic. In some remote Alaskan Villages, more than 70% of all adults died. Estimates of the final death toll worldwide vary widely, from 20 to 100 million, but were high enough that overall life expectancy was notably reduced.
The death toll exceeded that produced by World War I (WW I), which was ongoing at that time. In fact, 80% of deaths in the U.S. Army during WW I resulted from influenza and it is thought that the final collapse of the Germany army in 1918 may have been precipitated by widespread influenza in the troops. The surgeon general of the United States had expressed the hope that WW I would be the first war in which more U.S. soldiers died of war injuries than died of disease, but this hope was shattered by the influenza epidemic. The reasons for extreme virulence of the 1918 virus, and why healthy young people were more likely to die, remains a mystery, But the devastation caused by this virus raises continuing concern that a strain of influenza of equal virulence might appear and again cause immense suffering worldwide. Such concerns are heightened by the continual appearances of new strains of influenza virus and the fact that a strain of the virus epidemic in 1933 (the H1N1 strain) reappeared essentially unchanged 20 years later and caused a new epidemic. Thus, it is possible that the 1918 strain itself might reemerge. The pandemic of 1918 occurred before influenza virus could be isolated and it has not been possible to study the virus in the laboratory using modern tools. However, the sequences of the HA and NA gene of the 1918 virus have been obtained recently in a feat that demonstrates the power of modern molecular biology. Samples of preserved lung tissue taken at autopsy from two U.S. soldiers who died of influenza in 1918 were found to contain detectable influenza RNA, albeit in fragmented condition. Reverse transcriptase-polymerase chain reaction technology was used to obtain sequences from HA and NA that could be used to reconstructed the completed sequences of these genes. A third source of influenza RNA come from Alaskan victim of the 1918 influenza who had been buried in permafrost, and whose body was sufficiently well preserved that lung samples containing (fragmented) viral RNA were obtained. The sequences from these three victims were almost identical and showed that the virus belong to strain H1N1.
Asian flu, 1957-1958 [A (H2N2)]: It was caused about 70,000 deaths in the United States. First identified in China in late February 1957, the Asian flu spread to the United States by June 1957.
The Hong Kong Flu, 1968-1969 [A (H3N2)]: Viruses were first isolated in Hong Kong in July 1968. Widespread disease with increased excess mortality was observed in the United States during the winter of 1968–1969. Attack rates were highest (40%) among 10- to 14year-old children. Total influenza-associated excess mortality for this pandemic was estimated at 33,800 in the United States.
Prophylaxis:
Chemoprophylaxis: Influenza A viruses-but not B or C- are inhibited by amantadine, a primary amine, and rimantadine, a methylated derivative. Two anti-nucleic acid drugs, which inhibit viruses from budding from the cell surface, have now been licensed. They can inhibit influenza A and B viruses.
Influenza Vaccines: At present, immunization, rather than chemoprophylaxis, is the method of choice for preventing both influenza A and B. Even so, immunization poses a particular problem: every time a new strain of influenza A appears, the rapid production of large quantities of vaccine virus with required antigen characteristics, together with the need for routine tests of safety and efficacy, limits the amount of vaccine available. Most national authorities immunize about 10 percent of the population annually: these are two main categories of at-risk persons and it is most important that these individuals are vaccinated each year: the old (>75 years) or debilitated and those with chronic heart, respiratory, renal or endocrine disease; children; as well as people in closed institutions, such as residential homes for elderly, in which attack rates may be high. It is also considered for healthcare staff and police, who may need protection against wholesale sickness at times of major epidemics. Inactivated vaccines are prepared from appropriate strains of influenza A and B grown in the chick embryo allantoic cavity; the infected fluids are harvested, purified by ultracentrifugation, and inactivated with formalin or _-propiolactone. Most of the vaccines are either subunit preparations containing purified HA and NA or so called “split” vaccines that have been extracted with ether and detergent to reduce the side-effects of whole-virus vaccines. Apart from local erythema and soreness, sometimes with fever, these vaccines are generally very safe. “Live attenuated” vaccines are made by reassorting genes of viruses possessing the required HA and NA antigens with various laboratory-derived mutants selected previously for inability to growth at 37 C or for ability to grow only at low temperatures, for example 25 C (cold-adapted mutants). Both mutants are of diminished virulence for humans.
Conclusions:
Influenza A is a serious respiratory infection that places a heavy burden of disease on the global population. Many of the effects of influenza are hidden and the lack of effective treatments for the disease has tended to compound this situation. Furthermore, the full pathogenic profile in mammals of this highly labile and species-mobile pathogen remains incompletely understood. Experience during the 20th century tells us that we really do not know what to expect from the next influenza pandemic: will the next “lottery” result in a relatively benign phenotype or can we expect to see a truly virulent pantropic strain with the potential to kill millions?
During the past two decades, several widely held concepts concerning the epidemiology of influenza were demonstrated to be false. It was previously believed that influenza pandemics occurred at 10- to 14-year intervals, but it has been over 30 years since H3N2 viruses appeared. Furthermore, reclassification of influenza A viruses indicates that H1N1 viruses circulated from at least 1918 until 1957. Thus, it is now clear that influenza pandemics occur at unpredictable intervals. It was also believed that concurrent circulation of two different influenza A subtypes did not occur. However, H1N1 and H3N2 viruses have been circulating together since 1977. Finally, receptor specificity was believed to provide a barrier against human infection by avian influenza viruses that differ in this property from their human counterparts. This belief has been modified by the recently documented human infections by avian H5N1 and H9N2 viruses in Hong Kong.
References
1) Cinti S; Pandemic influenza: are we ready? Disaster Manag Response; 2005 Jul-Sep; 3(3):61-7.
2) James H. Strauss, Ellen G. Strauss; Viruses and Human Diseases; 2002; 147-156.
3) Leslie Collier, John Oxford, Jim Pipkin; Human Virology, second edition, 2000; 83-88.
4) Maria C. Zambon; Epidemiology and pathogenesis of influenza; Journal of Antimicrobial Chemotherapy (1999) 44, Topic B, 3-9.
5) Maurice R. Hilleman; Realities and enigmas of human viral influenza: pathogenesis, epidemiology and control; Vaccine 20 (2002) 3068–3087.
6) Peter Palese; Influenza: old and new threats; Nature Medicine; Nov, 2004, 10, S82 – S87.
7) Robert G. Webster, William J. Bean, Owen T. Gorman, Thomas M. Chambers and Yoshihiro Kawaoka; Evolution and Ecology of Influenza A Viruses; Microbiological Reviews, Mar. 1992, p. 152-179.