A HISTORY It began, as all things do, with a geology joke. We ranked candy based on their ability to elicit “joy” or “despair” and then worked in that ranking in various geological strata, both real and imagined. The strata, not the ranking. This started in 2006, and grew from there. At the beginning, we…
The Science Creative Quarterly
From archive
So Much Candy Data, Seriously
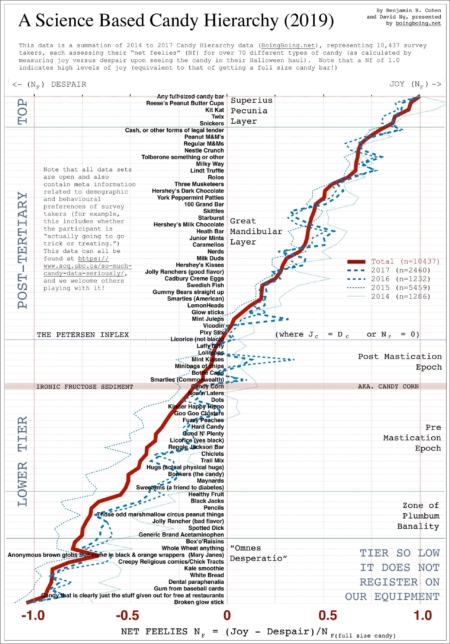
As promised, here is the candy hierarchy data for 2017. (Released Oct 25th @1:45pm PST. Will provide updated xlsx file on Oct 31st as well) . xlsx | csv | txt (d&t) | surveyQ pdf | n=2460 If you tag your work with #candyhierarchy2017 or #statscandy, I’ll do my best to aggregate efforts here (and also possibly at BoingBoing for a round up piece). Note that traditionally, we’ve plotted a figure based on a “Net Feelies” metric (=#JOY-#DESPAIR). We also have additional meta data affiliated with demographics, and some questions that link to other potential datasets (see actual survey for…
Advice for Potential Graduate Students – A Science Creative Quarterly Pin Up (No. 5)
– FROM THE ARCHIVES – (CLICK HERE FOR PIN-UP POSTER – pdf file ~85k – We suggest photocopying at 129% – LTR to 11×17) – – – We currently have room in the lab for more graduate students. But before you apply to this lab or any other, there are a few things to keep in mind. First, be realistic about graduate school. Graduate school in biology is not a sure path to success. Many students assume that they will eventually get a job just like their advisor’s. However, the average professor at a research university has three students at…
The 2020 Gairdner Award Winners: Picturing Science in the Classroom
Every year, the Gairdner Awards celebrate science and research excellence in the medical health areas. Since 1957, they have given out 395 awards – 95 of these recipients would go on to also win a Nobel Prize! In collaboration with the Canadian Society of Molecular Biosciences and the Michael Smith Laboratories at UBC, these materials were produced to provide a series of articles, comics, videos and accompanying lesson ideas to celebrate the science of a selection of the 2020 Canada Gairdner Awardees. This builds on the Gairdner Foundation’s partnership with CSMB and Michael Smith Laboratories at UBC, which began last…
How To Fly
“The knack of flying is learning how to throw yourself at the ground and miss” With these words, Douglas Adams helpfully explained concept of flying in his Hitchhiker’s Guide to the Galaxy. But the ground is really big, and, as the Tick so sagely noted, “Gravity is a harsh mistress.” So herein contained is my handy-dandy explanation of how you can impress your friends and family by throwing yourself at the ground and missing: Step one, throw yourself at the ground. Luckily, this is really easy thanks to gravity, which will pull you down to the ground at an acceleration…
A Serious Game on Gender Inequity and the Health Arena
It doesn’t take long to scan today’s headlines, and note the troubling incidents of #metoo, or hear word of research disparities that could potentially lead to life threatening outcomes for women. The reality is that even with the slow march of progress, there is still significant inequities in how the genders are treated in the health arena, if not society in general. This applies to both the medical research specifically (do treatments work better for men generally?), as well as the challenges that many women face in their career trajectories (how does gender affect careers?). Patriarchy, in a word, is…
Some COVID-19 Questions From a Curious and Concerned Seven Year Old
I got this letter the other day and it’s awesome! I thought I would try my best to answer these great COVID-19 questions. Thanks Alaina! 1. Where does the virus actually come from? Right now, the best answer is likely from one of these: Yup, a bat. But how it changed from a virus that infects bats to one that infects humans is still not really known. However, this sort of thing has happened before and the science word for it is zoonosis. This is where a disease which would normally only infect an animal (in this case a bat),…
Charity Wanjiku: Empowering with Power
Kenyan native Charity Wanjiku was first inspired to study architecture at the age of 10 when she saw a TV commercial for an insurance company where a businesswoman was presenting a model house to a boardroom of applauding men. Determined to pursue her passion — despite the fact that very few women enrolled in architecture courses — Wanjiku eventually graduated from the Jomo Kenyatta University of Agriculture and Technology with a Bachelor of Architecture degree. Later, she earned her Master’s degree in Project Management in Construction. It was at her first job where she discovered her interest in project management.…
Eugenia Duodu: Inspired, Elevated and Empowered
Dr. Eugenia Duodu earned an honours Bachelor of Science degree in Chemistry and Biology from the University of Toronto in 2010, and proceeded to study medicinal chemistry as a PhD student. Fueled by her passion to improve life for others, her research focused on providing effective treatments for human diseases such as cancer. Having grown up in a low-income community and experiencing numerous disparities to enriched science-based opportunities, Dr. Duodu sought out ways to share STEM with her community. It was at this time that she signed up as a part-time volunteer with Visions of Science Network for Learning Inc.…