GENETIC BASIS FOR FORCE-SENSITIVITY IN WOOKIEES
(This is the second paper on a special issue on Wookiee science. You can read the first here).
Annals of Praetachoral Mechanics. (2014). Vol 1. pp21-28 pdf download.
ABSTRACT
Midi-chlorians are sentient life forms, which live in endosymbiosis with all living cells (1). The number of midi-chlorians per cell has been found to correlate with influence in the force (2) and, as such, blood analysis has become common practice for the identification of force-sensitives, particularly by the Jedi Order. However, the genetic basis for increased midi-chlorian replication in force-sensitives is unknown. Here we show drivers to be mutations at both positions 263 and 312012 in Wookiee wookiee midi-chlorian DNA (mdDNA). We anticipate our findings to be a starting point for in vivo studies linking these mutations to force-sensitive phenotypes.
Key words: Force-sensitivity, midi-chlorian, single nucleotide variants
INTRODUCTION
The need for a powerful army has grown more pertinent in the wake of the rebellion against the Empire. Much research has been aimed at creating powerful fighters for the Sith (3). Sand People, Twi’lek, and Mon Calamari have all been considered, but Wookiees (Wookiee wookiee) are of particular interest due to their intelligence, large size, incredible strength, and unwavering loyalty (4). While powerful in their own right, Wookiees armed with the ability to harness the force have the potential be unstoppable soldiers. As the ability to harness the force has been correlated with midi-chlorian count in red blood cells (5), methods for increasing midi-chlorian count in Wookiee cells are of profound interest. However, and for as yet unknown reasons, all experiments that have aimed to increase expression of Wookiee specific midi-chlorians within Wookiee species have proved unsuccessful. More recently, evidence has demonstrated that force sensitive humans not only have higher numbers of midi-chlorians, but that several specific single nucleotide polymorphisms within the midi-chlorian genome may be responsible for hyper proliferation. Consequently, there have already been attempts to directly transplant force-sensitive-human midi-chlorians into Wookiee oocytes. However, these attempts have also failed, indicating Wookiee mdDNA and human mdDNA differ by genes essential for Wookiee cell proliferation (6). As a result, more advanced genetic techniques, and elucidation of midi-chlorian genome structure, are required to increase midi-chlorian count in Wookiees.
Here, we report sequence for both Human and Wookiee midi-chlorian genomes, and identify candidate mutations linked to Dark-side force-sensitivity. We then show characterization of synthesized artificial Wookiee mdDNA containing these candidate mutations. In particular, we determine effects on host-midi-chlorian replication by qPCR.
METHODS
Collection and Preparation of Blood Samples
mdDNA was isolated from blood samples as previously described (14). Briefly, blood cells were lysed by incubation with SDS and Protenase K, and then whole-cell lysates were subject to CsCl–bisbenzimide gradient centrifugation. CsCl–bisbenzimide gradients allow for separation of nuclear DNA from mdDNA based on A + T content (mdDNA is A+T rich). DNA quality was assessed by spectrophotometry (260 nm/280 nm and 260 nm/230 nm absorption ratios) and gel electrophoresis prior to library construction.
Midi-chlorian DNA Extraction
1ng input mdDNA was prepared using Nextera XT DNA Sample Preparation Kit (Illumina). Samples were barcoded with a 13bp unique index on each end to allow for analysis of 96 samples per flowcell, while maintaining coverage of >10000x per midi-chlorian genome. Libraries were sequenced with 2×150 bp reads on the Illumina JediSeq 2000 system.
Midi-chlorian Protein Extraction
Enriched midi-chlorians were lysed by adding 1ml Qiagen Native Lysis Buffer, and incubating on ice for 30 minutes with periodic gentle agitation.
Synthesis and Isolation of Midi-chlorian Genome
The Gibson et al. assembly method (9) was used to generate a synthetic midi-chlorian genome. It was important to minimize genome manipulation during the detergent and proteolytic enzyme treatments by suspending the cells in agarose blocks. Intact chromosomes were immobilized in the resulting cavern in the agarose that originally held the cell. Digested protein components, lipids, RNAs, and sheared genomic DNAs could then be removed by dialysis or electrophoresis from the immobilized intact genomic DNA. Whole, intact genomic DNA isolation was performed using a CHEF Mammalian Genomic DNA Plug Kit from Bio-Rad (7).
Isolation of midi-chlorians
Human and Wookiee blood samples were centrifuged at 600xg for 15 minutes to pellet cells. Cell pellets were resuspended in PBS to a final concentration of 1×108 cells/ml. Cell suspensions were lysed by forced movement through a 27G syringe needle, in the presence of a protease inhibitor cocktail (Roche, Germany). Crude cell lysate was incubated with 30ul anti-MATR microbeads for 60 minutes at 4°C. Treated lysate was loaded onto preequilibrated MACs column (Milyenyi Biotec). The column was placed in a MACS Separator (Miltenyi Biotec) and then midi-chlorians were washed and eluted according to Hornid-Do et al.’s method (8). Midi-chlorians were then pelleted and resuspended in MdIB (Midichlorian Incubation Buffer, Sigma-Aldrich).
Removal of original midi-chlorian genome
The native midi-chlorian genome was extracted under an inverted microscope (Nikon; model TMD). Cored Glass Tubes (Narishige; GD-1: I. D., 0.75 mm x O. D. 1 mm x length, 90 mm) and a Pressure Microinjector (Narishige; model IM-5B) were used with the added control of a Joystick Hydraulic Micromanipulator ((Narishige; system MO-204) to apply gentle suction and remove the native genome.
Insertion of synthesized midi-chlorian genome
CHEW5 cell line obtained from Dr. Darklord at Imperial University. CHEW5 in a 6-ml culture of SOB medium containing 17% fetal bovine serum and 0.5% glucose. Incubation was at 37°C until the medium pH was 6.2. Cells (5 to 50 × 107 cells/ml) were then spun in a centrifuge at 4575g for 15 min at 10°C. Cells were washed once [Tris 10 mM and NaCl 250 mM (pH 6.5)], resuspended with 200 ml of CaCl2 (0.1 M), and held on ice for 30 min. Synthetic midi-chlorian genome agarose plug was then added.
Real-time PCR
Primers and probes were designed using Primer 3, and checked for possible hairpin and self-dimer formation using IDT Oligo Analyzer 3.1. swBLAST was used to ensure specificity to the target of interest. Assays were validated, both in singleplex and multiplex, using dilution series of purified Wookiee mdDNA and purified Wookiee nuclear DNA (supplemental).
GAPDH Forward Primer: TCTCTGCTTCTGATGGCTCAAAC
GAPDH Reverse Primer: TGCTCTTCCGATCTGACAGCGACC
GAPDH Probe: 5’-FAM-TATTCGAGTAGC-MGBNFQ-3’
WOMDC Forward Primer: GGCTACCTATGAGGTCACTTTA
WOMDC Reverse Primer: TTCAGGTAGTAACACTGGGTA
WOMDC Probe: 5’-VIC-GGCTATCAACGT-MGBNFQ-3’
Real-time PCR was performed using the TaqMan® Fast Universal PCR Master Mix from Applied Biosystems (10ul of TaqMan® Fast Universal PCR Master Mix (2✕), No AmpErase® UNG, 0.5ul of 10uM GAPDH Probe, 0.8ul of 10uM GAPDH Forward Primer, 0.8ul of 10uM GAPDH Reverse Primer, 0.5ul of10uM WOMDC Probe, 0.8ul of 10uM WOMDC Forward Primer, 0.8ul of 10uM WOMDC Reverse Primer, 4.8ul of H2O, and 1ul of lysed culture). Thermocycling was carried out on a BioRad DNA Engine with Chromo4 Real-time PCR Detector (hot-start at 95 °C for 20 s, and 50 cycles of 1 s at 95 °C and 20 s at 60 °C).
RESULTS AND DISCUSSION
Single Nucleotide Variant Determination
Whole blood samples were obtained from 96 non-force-sensitive (NFS) Humans, 10 Sith, and 86 Wookiees. Specimens were collected as part of a research project approved by the Empire Force Agency Research Ethics Board (EFA REB). We sequenced the mdDNA isolated from each sample. NFS Human, and Wookiee midi-chlorian genomes were de-novo assembled using ABySS (12) with N50 sizes ranging from 34624 to 50348. Single nucleotide polymorphisms (SNPs) within the NFS Human subset were identified using SAMtools (supplementary Figure 1).
Sith mdDNA reads were aligned to the NFS Human draft genome using Burrows-Wheeler Aligner version 0.5.4 (13). Screening for SNPs by SAMtools followed by subtraction of known SNPs from the NSF human data set yielded four single nucleotide variants (SNVs) at positions: 257 (A/G), 35768 (T/G), 274491 (G/T), and 289002 (T/G).
We then used Projector, a pair hidden Markov model-based program, to identify homologues to the Sith midi-chlorian genes containing SNVs in the Wookiee midi-chlorian genome (11). Candidate SNVs positions in the Wookiee midi-chlorian genome were then identified by visual inspection of gene homologs: 263 (A/G), 35769 (G/T), 266724 (A/G), and 312012 (C/T).
Midi-chlorian Synthetic Genome Synthesis
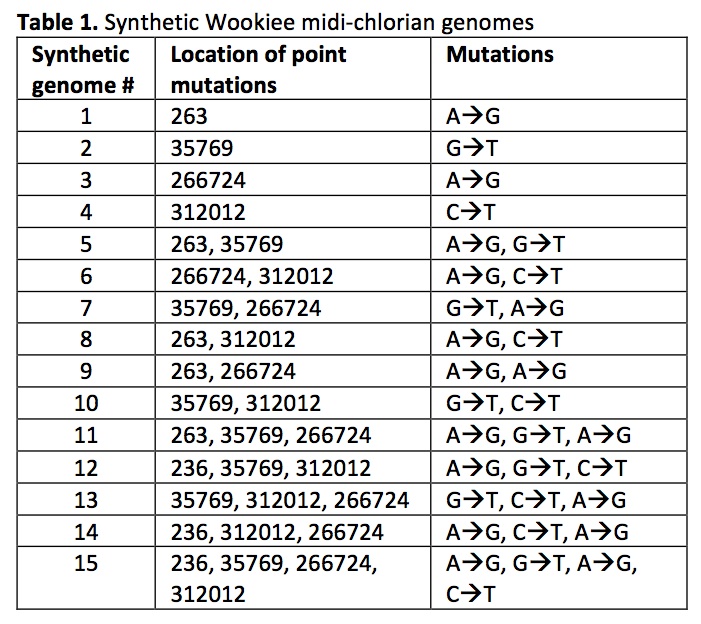
Using Gibson et al.’s technique for chemical genomic synthesis (7), we created 15 synthetic Wookiee midi-chlorian genomes containing mutations at the positions of the candidate SNVs identified (Table 1). Synthetic genomes were prepared for storage and transformation using CHEF Mammalian Genomic DNA Plug Kit from Bio-Rad, as in Gibson et al. (2009) (7).
CHEW5 Mutant Creation
Midi-chlorians were extracted from human and Wookiee blood, using Hornig-Do et al.’s magnetic field method, along with MACs Column and Separator from Miltenyi Biotec (8). Whole midichlorian extract was 99% pure, as determined through blotting of crude cell lysate and enriched midi-chlorians using probes for β-actin (cytoskeleton), GAPDH (cytosol), Rab4 (Golgi apparatus), Golgin-97 (endosome), and HanSo (midi-chlorian).
The native Midi-chlorian genome was removed under an inverted microscope using a microsyringe, leaving the wookie midi-chlorian intact. Midi-chlorian synthetic genomes were transferred into genome-free Wookie Midi-chlorians using the Lartigue et al. method (7), involving treatment of midi-chlorians with CaCl2 and then heat shock in the presence of synthetic genome agarose plugs. To ensure only midi-chlorians containing a single genome were carried through to subsequent steps, suspensions were incubated with a FISH probe specific to a repetitive region of the Wookiee midi-chlorian genome (6) and then FACS was used to sort midi-chlorians: negligible fluorescence intensity indicating that the midi-chlorian did not successfully take up the DNA, one arbitrary fluorescence unit indicating that the midi-chlorian took up one copy of the mdDNA, and two arbitrary fluorescence units indicating that the midichlorian took up two copies of the mdDNA etc. (Supplementary Methods). One midi-chlorian per cell was microinjected into CHEW5 cells, and presence was verified by optical microscopy.
Determination of Midi-chlorian Production by qPCR
Single CHEW5 cells containing midi-chlorians with synthetic genomes (SG), and control CHEW5 cells, were placed into wells of a 96-well plate and were cultured as described previously (6). Five replicates of each were seeded. At 6, 12, 24, 48, 72, and 96 hours after seeding, 2ul of each culture was extracted, lysed (Cell-md Direct Kit, Darth Distributors), and used as template in a two-colour qPCR assay. An assay specific to the constant region of Wookiee mdDNA (WOMDC) was used to assess quantity of midi-chlorians (VIC channel). To account for differences in cell proliferation among cultures, an assay for GAPDH, a housekeeping gene on Wookiee nuclear DNA, was used (FAM channel). The difference in cycle threshold values (ΔCT) between WOMDC and GAPDH assays was used to compare midi-chlorian replication (Figure 1).
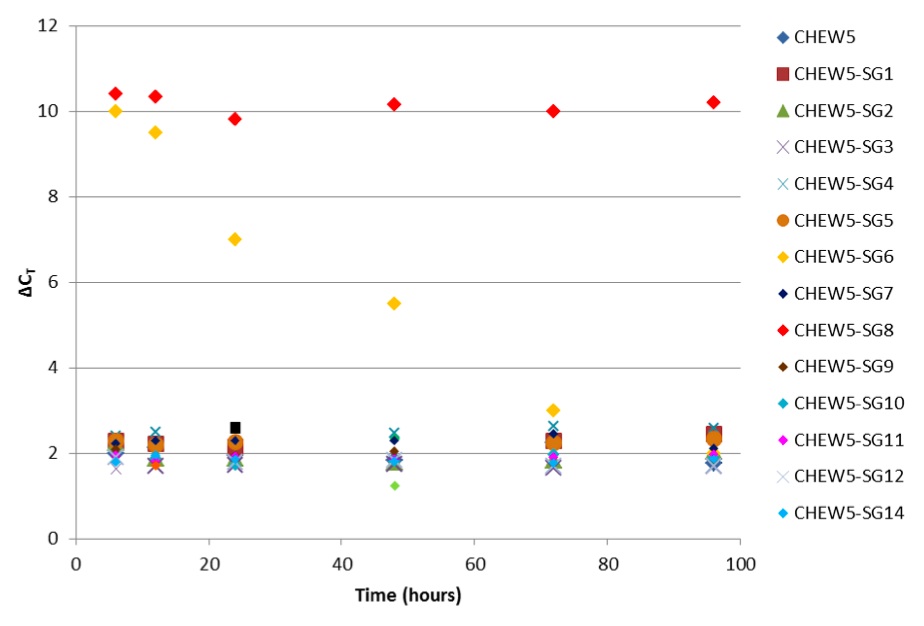
Figure 1. (Click to Enlarge): Difference in cycle threshold values (ΔCT) between WOMDC and GAPDH assays over 96 hours.
CHEW5-SG13 and CHEW5-SG15 mutants were not viable. Of the remaining 13 CHEW5 mutants, 10 did not significantly differ from normal CHEW5 in their midi-chlorian replication behaviour (p= 0.67, t test). Both CHEW5-SG6 cells and CHEW5-SG8 cells demonstrated high per-cell midichlorian counts, but CHEW5-SG6 mutants appeared unstable, as demonstrated by a consistently decreasing ΔCT over the six time points.
Growth curves for CHEW5-SG8 and CHEW5 cells were obtained by OD600 (Supplementary Figure 4). The presence of the midi-chlorian containing SG8 in the CHEW5 cells does not appear to affect CHEW5 proliferation.
Midi-chlorians were extracted (see Methods) from each CHEW5-SG8 culture, lysed, and the total proteomes were analyzed on a 2 dimensional gel (Figure 2). The boxes highlight a single protein difference between the CHEW5 and CHEW5-SG8 proteomes. As expected this protein corresponds to one found in Sith midi-chlorian.
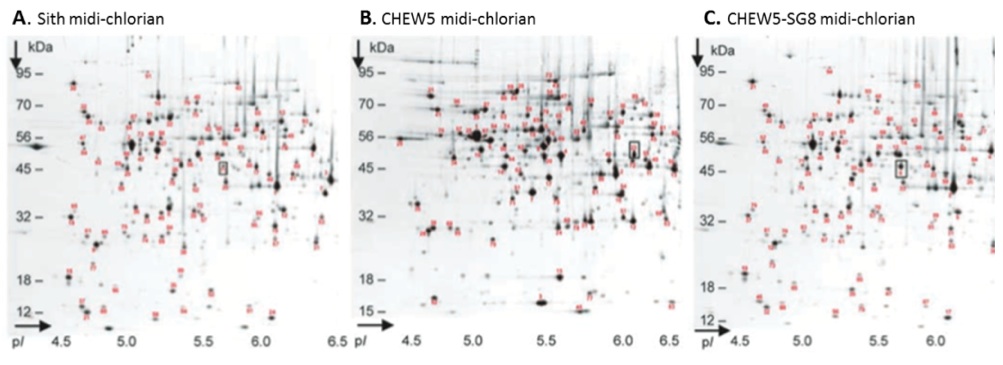
Figure 2 (Click to Enlarge). Midi-chlorian proteomic analysis. Two-dimensional gels were run using midi-chlorian lysates from (A) Sith cells (B) CHEW5 cells, and (C) CHEW5-SG8 cells. Standard conditions were used for the separation of protein spots in the first dimension on immobilized pH gradient (IPG) strips (pH range 4 to 7) and in the second, SDS-PAGE, dimension (molecular mass 8 to 200 kD) (10). Gels were stained with Coomassie brilliant blue G-250.
CONCLUSIONS
Four SNVs linked to dark-side force sensitivity were determined through sequence alignment of NFS Human and Sith mdDNA. Wookiee genomes containing mutations homologous to the identified human SNVs were synthesized and then inserted into midi-chlorians. Midi-chlorians containing each synthetic genome were inserted into CHEW5 cells. Because of the existing, well-documented link between force-sensitivity and increased midi-chlorian count (2) , qPCR assays were used to identify CHEW5 mutants producing high levels of midi-chlorians. The CHEW5-SG8 mutant, containing mutations at positions 263, and 312012, was found to steadily produce high levels of midi-chlorians. Analysis of the proteome of CHEW5-SG8 midi-chlorians revealed one protein also found in Sith midi-chlorians. To definitively determine that midi-chlorian SG8 imparts force-sensitivity to its host, in vivo experiments are required. Our group has begun preparations for a trial involving transplantation of midi-chlorians containing SG8 into Wookiee oocytes, and in vitro fertilization of these oocytes in to female Wookiees.
LITERATURE CITED
1. Baaka C (2012) in The Wookieepedia Volume 312: Midichondrians as Sentient Life, eds. Ng D, Smith M (Springer, Heidelberg), pp 403–420.
2. Baaka C (2012) in The Wookieepedia Volume 312: Midichondrian Count in Relation to Force Sensitivity, eds. Ng D, Smith M (Springer, Heidelberg), pp 1106–1158.
3. Plageuous D, Nemisis A (2424) The Clone Wars diaries: Attempt to create evil life. Proc Natl Acad Sci EARTH 966:10802-10831.
4. Skywalker L (1999) Physical and social characteristics of Wookiees: a field study. Gal Anal Biol 88:302-333.
5. Calrisian L (2008) in the jedi handbook on midi-chlorian count, ed Kanobe OBO (Springer, Heidelberg) 2:30-40.
6. Sotired I, Falcon M, Iwannagotosleep PLS (2011) Wookiee mdDNA and human mdDNA differ by genes essential for Wookiee cell proliferation, Science 172: 3076-3083.
7. Lartigue C, Glass JI, Alperovich N, Pieper R, Parmar PP, Hutchison III CA, Smith HO, Venter JC (2007) Genome transplantation in bacteria: changing one species to another, Science 317(632); 632-638.
8. Hornig-Do HT, Günther G, Bust M, Lehnartz P, Bosio A, Wiesner RJ (2009) Isolation of functional pure mitochondria by superparamagnetic microbeads, Anal Biochem 389(1): 1-5.
9. Gibson DG, et al. (2010) Creation of a bacterial cell controlled by a chemically synthesized genome. Science 329(52);52-56.
10. Outofideas IAM, (2013) in Intergalactic species midi-chondrial proteome project: humanoids, ed Dorky I, (Silly University Press) pp 300078-487699
11. Meyer IM, Durbin R (2004) Gene structure conservation aids similarity based gene prediction. Nucleic Acid Res. 32(2); 776-783.
12. Simpson JT (2009) ABySS: A parallel assembler for short read sequence data. Genome Res. 19(6): 1117–1123.
13. Li H, Durbin R (2010) Fast and accurate long-read alignment with Burrows-Wheeler Transform. Bioinformatics 26(5):589-95
14. Lang F, and Burger G (2007) Purification of mitochondrial and plastid DNA. Nature Protocols 2(3): 652-660.