TRANSCRIPTOME DIVERGENCE BETWEEN COMMON AND CAVE-ADAPTED WOOKIEES
– – –
Annals of Praetachoral Mechanics. (2014). Vol 1. pp7-19 pdf download.
ABSTRACT:
Cave-adapted organisms commonly evolve convergent morphological, physiological and behavioral traits. The rare cave-adapted wookie species, Wookiee troglodytus, presents an opportunity to characterize underlying evolutionary mechanisms driving phenotypic expression in a complex sentient humanoid. The cave wookie presents several features associated with adaptation to subterranean habitats: pigment-less hair, reduced visual acuity, overall smaller stature, elongated limbs and decreased aggression. Uncovering the evolutionary processes directing the complex morphological and behavioral traits requires genome-level analysis. To unpack these complex traits, complete nuclear transcriptomes and enrichment profiles were assessed for common wookie populations and the troglomorphic species. Common and troglomorph wookie representative transcriptomes were uniquely divergent in genes and expression levels associated with pigmentation, vision, adrenal gland function, and metabolism.
Key words: Fst, transcriptome, troglomorph, troglobite, sentient
INTRODUCTION
Organisms of widely different evolutionary histories frequently present convergent morphology associated with adaptation to life in caves. “Cave,” as a habitat, is a classification derived from diverse subtypes but shared characteristics of no light, frequently low nutrient levels and high humidity are thought to exert varied and strong selective pressure for subterranean-adaptive features (Romero 2011). Some of these features include: reduced pigmentation, reduced visual-sense apparatus paired to enhanced non-visual sensory systems, decreased or enlarged body size, elongated appendages, reduced aggression and alarm-response (Romero 2011). A recently described species of cave-adapted wookiees present interesting convergent features that correspond with their almost exclusively subterranean life history: pigment-less hair, poor visual acuity, small stature, and extreme passivity. The species is comprised of two known populations that went undetected until 2002 when biospeleotic surveys were prompted after rare sightings of albino wookiees (Dubois and Sinclair 1990, Tong 1993). The survey, conducted by Sphinx et al. (2003), uncovered two extensive cave networks in the northwest Wawaatt archipelago, each resident to populations of pigment-less wookiees. Spikens (2012) classified the two populations as a single new species based on mitochondrial DNA phylogenetic analysis and morphology. In observed specimen, morphological analysis demonstrated: smaller stature (relative to common wookiees), skin and hair absent of pigment, and reduced visual sense (Spickens 2012). Phylogenetic analysis of cytB and 12S mtDNA placed individuals from the two northwest Wawatt cave populations in a single clade sister to a monophyletic clade of individuals from widely distributed populations of W. wookiee. (Spickens 2012). Spickens (2012) described the species, Wookiee troglodytus.
While suites of troglomorphic traits are common, degree of morphological, biochemical and behavioral adaptation varies amongst cave-adapted species because of the independent nature of each adaptive event (Romero 2011). Cave-adapted species display convergent phenotypes but the underlying biological processes may be unique between organisms of different phylogenetic history and in different cave habitats. Additionally, pigment, vision, body arrangement and behavior are complex traits controlled by many simultaneously operating genes and molecular pathways. Genomics based approaches have proven useful in elucidating genetic differences between related taxa presenting complex phenotypic differences as the result of adapting to novel habitats (Gross et al. 2013, Kozak et al 2013).
Gross et al. (2013) identified regions of sequence divergence between transcriptomes of subterranean and surface morphotypes of the Mexican blind cavefish, Astyanax mexicanus. They chose to integrate separately sequenced surface and cave-adapted A. mexicanus transcriptomes into a combined data-set which proved useful for evaluating conserved variation amongst morphotypes. RNAseq data-mining allowed the researchers to identify genes expressed exclusively in either morphotype. Their results showed decreased expression of multiple vision system genes and increased expression of multiple metabolic genes in troglomorph cavefish compared to surface-dwelling cavefish. Genetic mechanisms for physiological adaptation to fresh-water and salt-water habitat between two species of Killifish (Lucaina parva and Lucaina goodyi) and between two interspecifc populations (Lucaina parva) were the subject of Kozak et al. paper (2013). The researchers identified gene regions of high differentiation between fresh and salt-water species and used GO enrichment analyses to uncover the genetic component enabling populations to inhabit different niches. They found differentiated gene-expression in multiple osmoregulation genes between and within species. These experiments demonstrate that comparative transcriptomics can be useful in identifying genetic evolution between phenotypically divergent related taxa.
The diverse evolutionary history of troglomorphs requires a more complete understanding the breadth of genetic evolution among cave organisms. We investigated the underlying genome evolution in W. wookiee and W. troglodytus, as the mechanisms driving phenotypic divergence in cave and terrestrial taxa had yet to be explored in a sentient organism. Given the morphological and behavioral adaptations reported by Spickens (2012), we hypothesized that genes involved in pigment, vision and adrenal system pathways would highly divergent between species. We assembled and analyzed complete transcriptomes from pooled and individually genotyped male and female W. wookiee from four wide-dispersed populations and male and female W. troglodytus from each cave population. Quantifying genome-wide sequence variation between species and populations enabled us to identify genes likely involved in adaptation to caves.
Genome-wide analysis of transcribed genes will further the understanding of divergence between W. wookiee and W. troglodytus and identify what genes are differentially expressed, connecting genotype and expression to phenotype. It will also enable further comparisons between troglomorph transcriptomes of different lineages and complexities, and continue to uncover patterns of genetic evolution in cave habitats.
METHODS
Source Populations
We sampled specimen from each of the two populations of W. troglodytus and four geographically distinct populations of W. wookiee. Each sample population of W. wookiee was a minimum of 1500 km and a maximum of 3000 km from an adjacent sampled population.
Transcriptome
For each population we sampled healthy tissue from five male and five female individuals. Total RNA was extracted using Trizol (in supplementary material) and purified using RNeasy mini kits (Qiagen, Valencia, CA). Male and female samples were pooled by population with an equal proportion of total RNA from each sampled individual (concentrations determined by qPCR).
Pooled samples were sent to Unigens (New York, USA) for library preparation and sequencing on an Illumina HiSeq2000.
Transcriptome Assembly
Illumina sequences were assembled de novo using Velvet (Zerbino and Birney 2008) after being trimmed (removing the first and last 20 bp) using the FastX trimmer. The assemblies were created from all Wookiee spp. sequences using the paired-end option, an insert size of 200 bp, and we tried several of kmer values. The best assembly was under kmer of 20, which generated contigs over 100 bp as follows: 121,636 contigs for troglomorph population 1 (T1) wookiees, 100,030 contigs for troglomorph population 2 (T2) wookiees and 94,142 contigs for the common-wookiees (CW). We found that 92% of the T1 contigs, and 89% of the T2 contigs were represented in the CW assembly. We created our reference based on the CW assembly with a total of 29,881 contigs, combining 2243 contigs from the T1 and T2 assemblies (all unique contigs over 300 bp) with CW contigs that had blast matches to any of the following: (1) related-wookiee proteomes, or (2) target genes of interest (pigmentation, vision and adrenal gland genes). The software suite Galaxy (Giardine et al. 2005) was used to filter results from blastX searches against the uniprot proteomes and retain only contigs that had only 1–3 protein matches per species (blast score ≥ 100). This was done to avoid contigs that were misassembled from multiple genes.
SNP detection and Fst calculations
Each wookiee population was aligned separately to our constructed reference using BWA and its paired-end function from (Li and Durbin 2009). Alignments were then processed with samtools (Li et al. 2009). Single nucleotide polymorphism (SNP) detection and Fst calculation was performed with the software Popoolation2, which calculates population genetic metrics from pooled samples (Kofler et al. 2011). Allele frequencies and Fst between populations were calculated with a minimum coverage of 10 reads per population in order to exclude SNPs from low expressed transcripts. We used a pool size of 5, and a sliding window of 20 bp. The mean number of reads per population for contigs with SNPs was 38.09 in a range of 26.12–51.16. This is an initial step to identify candidate SNPs.
To validate allele frequencies from our pooled analysis, we genotyped individuals from each population at 3485 SNPs using an Illumina Infinium Bead Chip custom designed for Wookiee (containing 1000 candidate SNPs segregating between species, 1300 SNPs segregating between W. wookiee populations and 965 SNPs segregating between W. troglodytus). Bead design was adapted from Kozak et al. (2013) and genotyping protocol can be found in Supporting Information. We genotyped 16 W. wookiee and 12 W. troglodytus using genomic DNA extracted from cheek cells (preserved in ethanol) following the Gentra PureGene protocol (Qiagen).
Inter and Intraspecies Analyses
To look at divergence across troglomorphic adaptations we compared Fst analyses between CW, T1 and T2 wookiees. Fst outlier loci were identified in two sets of contrasts. First, we performed an outlier analysis between interspecific populations to identify loci that evolve quickly between population pair, but are not relevant to subterranean evolution. In CW we compared the Fst in sliding windows between two most geographically distant populations. The same comparison was made between T1 and T2. Second, we compared pooled-common and T1 or T2 pairs. We excluded from consideration as candidate troglomorphic adaptations the outliers that overlapped with those from the geographically distinct cave-dwelling wookiees. In this way, Fst outliers between cave-dwelling and common wookiee pairs should capture adaptive variation across niche-habitats. We then focused on SNPs that were outliers in cave-common pairs in order to detect loci that repeatedly evolve in different light, nutrient and humidity environments.
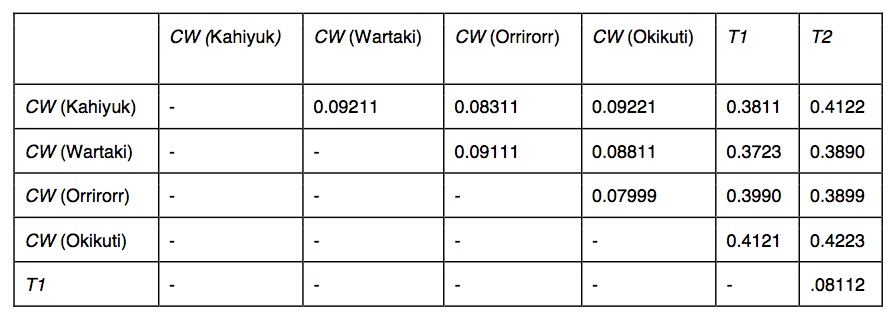
Table 1. (click to enlarge) Genome-wide Fst estimated between all Wookiee populations across all 38,126 SNP intervals.
For individual genotypes, we also performed Fst estimation with Infinium bead genotype data. The hierfstat package in R was used to calculate Fst at all possible SNPs with segregating alleles for each of the species/population comparisons (Goudet 2005). The SNP bead probe sequences were aligned with the transcriptome reference using blastn. Genomic SNPs were paired with the corresponding 50 bp window from the pooled analysis and we compared Fst values
from individual genotypes to our pooled data. With the Infinium genotype data, we performed two GO enrichment analyses on outlier loci: (1) between wookiee species and (2) between all cave-dwelling and all common wookiee individuals.
The Fst outliers were identified based on empirical distributions of divergent loci (Beaumont and Balding 2004; Narum and Hess 2011). This approach has been used in several model organisms (Kolaczkowski et al. 2011, Turner et al. 2010, Akey et al. 2010) and in humans (Akey et al. 2002), to identify putative targets of selection.
RESULTS
Transcriptome divergence between cave and terrestrial populations
We identified 205,604 SNPs that were segregating among our six populations. Fst was calculated over 38,126 intervals (50 bp in length) with at least one segregating SNP from 7366 contigs. These contigs were mapped to 3454 genes with functional annotations. Distribution and average Fst values for population/species pairs are shown in Table 1.
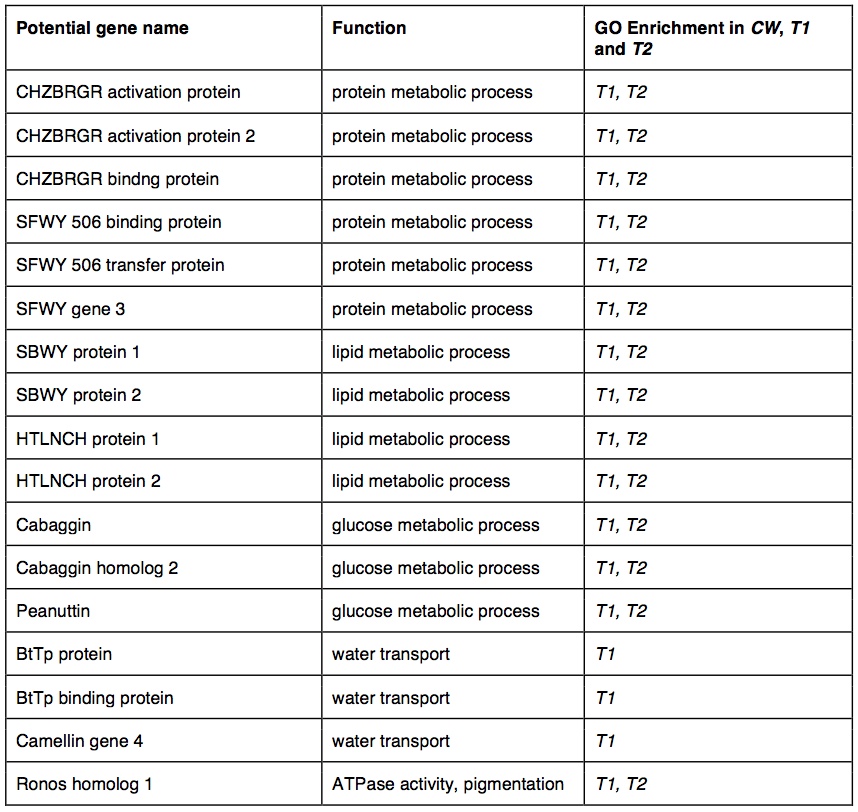
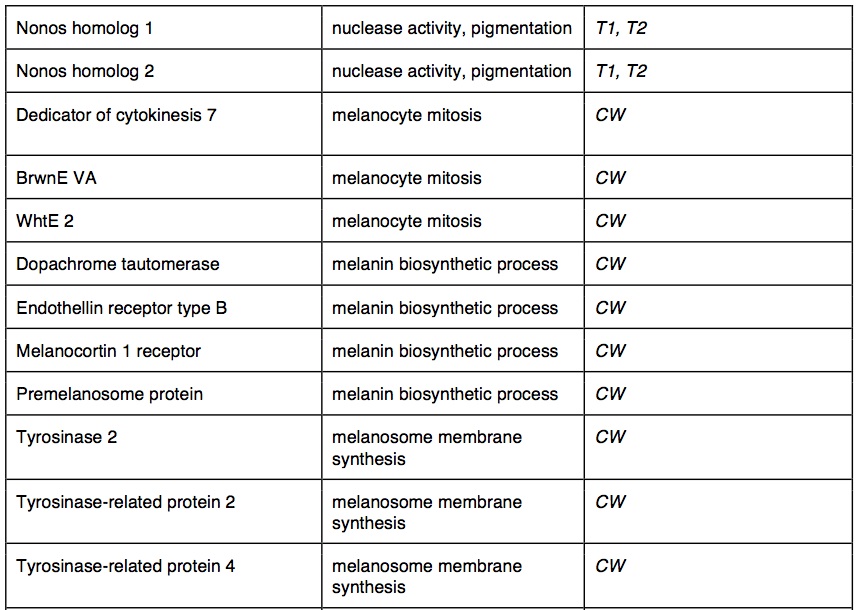
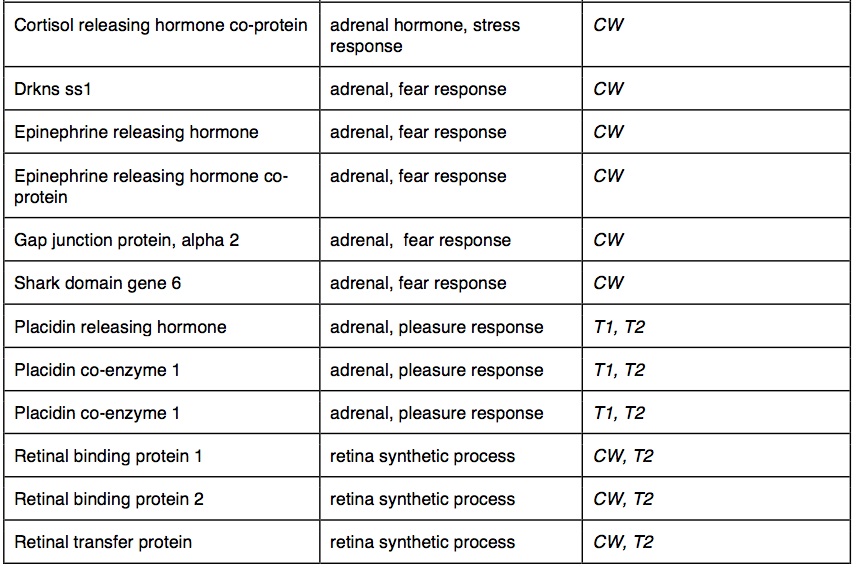
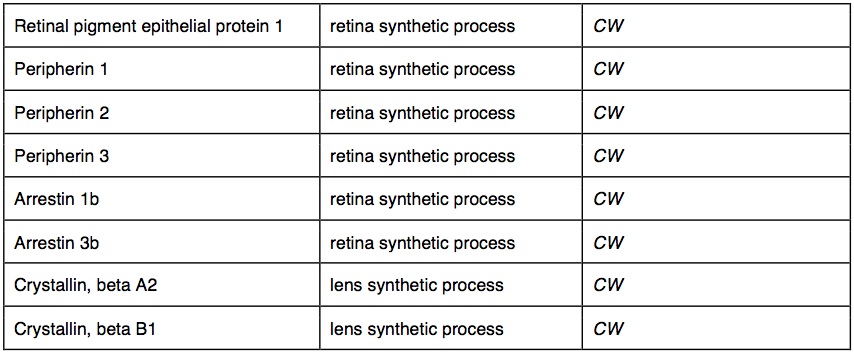
Between species, 2360 of the 50 bp windows had one or more SNPs that were completely fixed and identified as outliers between species (top 5% of Fst distribution: average Fst = 1). These outlier SNPs were in 1756 contigs that mapped to 1239 genes with GO annotations. The outliers had significant GO enrichment in 150 biological processes and 90 molecular functions. Many of the GO-enriched categories were related to functions predicted to be under selection in cave-adapted organisms. Several of these potential genes were involved in metabolic activity (30 genes), adrenal activity (20 genes), pigment synthesis (20 genes) and visual sense (17 genes). We identified outlier water transport genes. In cave wookiees metabolic activity was significantly enriched (three biological processes). We identified multiple retina and lens specific genes of significant enrichment in CW and absent in T1 and T2. Pigment synthesis was significantly enriched in CW (three biological processes, two molecular functions). T1 and T2 transcriptomes revealed several significantly down regulated genes involved in adrenal hormone synthesis (three biological processes). Outlier genes and enriched taxa are listed in table 2.
Divergence between caves populations
To determine which loci evolve quickly between populations due to processes other than adaptation to light and humidity, we compared divergence between our two cave-dwelling populations.
Between our T1 and T2 populations, we identified 402 outlier SNP windows (Fst ≥ 0.321). We found 53 enriched biological processes, 17 cellular components, and 12 molecular functions. In T1 we found unique down-regulation of three genes for retinal synthesis.
Divergence between geographically distinct terrestrial populations
To determine which loci evolve quickly between populations due to processes other than adaptation to caves, we compared divergence between our two most geographically distant W. wookiee populations. We identified 2402 outlier SNP windows (Fst ≥ 0.421). We found 73 enriched biological processes, 13 cellular components, and 16 molecular functions. We found enrichment of two potential epinephrine regulatory biological processes (coupled ATPase activity, protein serine/threonine kinase activity) and one reproductive process (C21-steroid hormone metabolic process). These outliers were removed to control for variation from genetic drift and factors unrelated to habitat adaptation.
Pooled Sequence Divergence versus Individual Sequence Divergence
We found similar levels of divergence between species when comparing W. wookiee and W. troglodytus individuals genotyped on Infinium bead chips (16 W. wookiee, 12 W. troglodytus). Average Fst across 3485 SNPs was 0.386 +/- 0.006, similar to the mean Fst of 0.38 from the pooled analysis (t-test assuming unequal variance: t4, 881 = 0.33, P = 0.74). Between species Fst estimates showed significant positive correlation between pooled and Infinium data (SNPs aligned within 50 bp, Pearson r = 0.49, P < 0.0001, N = 299). Pooled Fst estimates were not significantly higher than individual genotype data. Of the outlier loci between species identified in the pooled analysis 88% also showed high sequence divergence in the genotypes.
Table 2. (Click to enlarge) Summary of gene outliers and GO enrichment between common wookiee (CW) and troglomorph wookiee populations (T1 and T2)
CW genotype GO enrichment analysis on Infinium SNP data was compared to pooled sample CW enrichment analysis. Infinium SNP data recovered enrichment of two epinephrine regulatory biological processes and one reproductive process. One thousand Fst outliers (0.3833 ≤ Fst ≤ 0.4016) were identified between CW, T1 and T2 populations using Infinium SNP data that corresponded to 980 genes enriched in metabolic, adrenal, pigment, and vision biological processes. Water transport genes were not enriched in genotype GO analysis.
DISCUSSION
W. troglodytus and W. wookiee live in highly contrasting environments subject to unique selective pressures. These differences were reflected in the genomic data. We discovered many contigs that contained highly differentiated SNPs associated with multiple biological processes that correspond with phenotype. Metabolic, adrenal activity, pigmentation, vision and water transport genes were highly differentiated between transcriptomes of common and cave-wookiees. Genetic outliers identified between species of Wookiee were not identified in interspecies comparisons, thus we feel confident the differences reflect shared, cave- or surface-evolution over variation due to geographic distance and genetic drift.
We suspected some visual genes may be compromised in W. troglodytus, but the quantity (17 genes) was unexpected. Troglomorph wookiees have poor visual acuity compared to surface species, but the acuity is reduced, not non-functional. Complete eye loss is common in troglomorphic species but it has not yet been observed in sentient troglomorphs (Protas and Jeffery 2012). Where eyes are absent, loss has been shown to result from initial eye development halted when retina and retina pigment epithelia (RPE) cells undergo apoptotic cell death (Protas and Jeffery 2012). We are surprised that at least one RPE gene is showing down-regulation across troglomorph wookiees while visual function is still possible. We were also surprised to find two retinal binding proteins and one retinal transfer protein expressed in common wookiees and one population of cave wookiees, and down-regulated expression in one population of troglomorph wookiees.
Multiple metabolic process genes were divergent between W. wookiee and W. troglodytus. Astyanax cave and surface morphotypes showed enriched GO for metabolic processes that were in concert with a physically reduced pineal gland (Gross et al. 2013, Omura 1975). Predictably, outlier genes identified between cave and surface morphotypes were involved in lipid and protein metabolism (Gross et al. 2013). Gross et al. (2013) hypothesized that reduced nutrient and oxygen supply to subterranean habitats may impose selective pressure upon cave morphotypes to evolve more efficient metabolism, which is physically manifested in a smaller pineal gland. Outlier genes between W. wookiee and W. troglodytus were identified in protein, lipid metabolism, and glucose metabolism. We suspect that the enrichment of these genes across W. troglodytus populations represents selective pressure for efficiency in this species as well. However, physical abnormality in the pineal gland has not been observed in troglomorph wookiees.
This analysis revealed, across troglomorph wookiees, three outliers associated with water transport in pooled-sample transcriptomes that were not recovered in individual genotype enrichment analyses. Camellins and BtTp proteins play a crucial role regulating cellular osmotic stress and hypohydraemic metabolism. We did not expect evolution in troglomorph wookiees to be under strong selective pressure for efficient water transport; the cave network habitats are, on average, more humid than surface habitats on Kashykk. It may be that potable water is limited in these caves or that another factor is exerting selective pressure on cellular water transport.
Reductions in pigment gene expression were not surprising. W. troglodytus are readily identifiable by the absence of color in their hair and skin. Our results identify underlying mechanisms of one of the most apparent phenotypic differences among troglomorph and common wookiees. Three biological processes associated with pigmentation were enriched in W. wookiee: melanin biosynthesis, melanocyte mitosis, and melanosome membrane biosynthesis. One of the genes identified in W. wookiee most strongly expressed is BrwnE VA. In albino W. wookiee, the gene is mutated and fails to facilitate chromatin coiling within melanocytes (Peterson and Peterson 2001). Peterson and Peterson (2001) identified non-function tyrosinase genes in melanocytes of albino W. wookiee and Ewok ewok. Melanocytes, where tyrosinase production was limited, formed incomplete membranes. Crucial components of pigment synthesis are compromised in W. troglodytus. We suspect pigmentation genes are mutating under relaxed selective constraints for pigment synthesis among troglomorph wookiees.
Some of the more curious observations of W. troglodytus, recorded by Sphinx et al. 2003 and Spickens (2012), are of their behavior. Fair-minded but excitable, common wookiees interact aggressively to resolve disputes. Wookiee on wookiee violence is less frequent, but not unheard of, and they cooperate to combat threat. Increased cortisol and epinephrine production has been demonstrated in conflicting common wookiees (Grey and Green 1986). Sphinx et al. (2003) and Spickens (2012) noted arguments among troglomorph wookiees conspicuously absent and observed zero incidences of aggressive posturing. Common wookiees are reported to engage in limited social grooming, most often displayed between very young siblings and cousins (Spickens 1979). Spickens (2012) recorded W. troglodytus engaging in social grooming for as many as 8 hours a day. Grooming behavior was observed across ages and genders (Spickens 2012). In common wookiees, social grooming stimulates the release of placidin, a mildly tranquilizing hormone (Grey et al. 1984). Adrenal hormone synthetic processes associated with placidin, cortisol and epinephrine were highly variable between cave-adapted and common wookiees. Among the outliers, cortisol and epinephrine synthetic pathways plus associated proteins were enriched in W. wookiee. Placidin genes were significantly enriched in W. troglodytus. Variation in gene expression across these hormone synthesis processes likely accounts for the unique behavioral adaptations between wookiee species. Increased grooming behavior among troglomorph wookiees may be explained by selective pressure to maintain good hygiene in a humid and dark environment. Alternatively, tactile sense may be elevated in troglomorph wookiees, as non-visual senses are often heightened in the absence of light (Romero 2011). In either case, the behavior is likely reinforced by the pleasant feeling associated with placidin production (Spickens 1979).
CONCLUSIONS
In this report, we present the results of comparative transcriptome analysis between surface and subterranean-adapted wookiee species, W. wookiee and W. troglodytus. Our data uncovered divergence in gene expression between the species that is consistent with observed morphological and behavioral differences. Many of the highly variable genes identified in our research have been indicated as important factors other cave-adapted organisms (Reviewed in Protas and Jeffery 2012). Areas of further research should begin with deeper investigation of vision and endocrine system development between wookiee species and between populations W. troglodytus.
ACKNOWLEDGEMENTS
The authors thank the staff of Unigens for providing ample support in transcriptome analysis (New York City, New York); Q Viceroy assisted with bioinformatics; P Spickens and J Sphinx provided observations and discussion of in situ wookiee populations. The transcriptome sequencing work was approved by the University of British Columbia and was funded by the Universal Humanoid Research Foundation Awards (ZX0-1222 to PCQ). B Wang, T Cheeke and three anonymous reviewers provided helpful comments on the manuscript.
LITERATURE CITED
Akey JM, Zhang G, Zhang K, Jin L, and Shriver MD. 2002. Interrogating a high-density SNP map for signatures of natural selection. Genome Res. 12:1805–1814.
Akey JM, Ruhe AL, Akey DT, Wong AK, Connelly CF, Madeoy J, Nicholas TJ and Neff MW. 2010. Tracking footprints of artificial selection in the dog genome. Proceedings of the National Academy of Sciences USA 107:1160–1165.
Beaumont,M. A., and D. J. Balding. 2004. Identifying adaptive genetic divergence among populations from genome scans. Mol. Ecol. 13:969–980.
Dubois P and Sinclair V. 1990. White wookiee? Abino wookiee sighted in northwest Wawaat. Biological Transactions 10: 12-14.
Grey P, Chan V, Truong K, Lian C. 1983. Placidin: structure and function. Journal of Sentient Endocrinology 281: 64-71.
Grey P and Green V. 1986. In-combat W. wookiee hormone secretion. Journal of Sentient Endocrinology 340: 12-17.
Kofler, R., R. V. Pandey, and C. Schlotterer. 2011. PoPoolation2: identifying differentiation between populations using sequencing of pooled DNA samples (Pool-Seq). Bioinformatics 27:3435–3436.
Kolaczkowski B, AD Kern, AK Holloway, and D Begun. 2011. Genomic differentiation between temperate and tropical Australian populations of Drosophila melanogaster. Genetics 187:245–260.
Kozak GM Brennan RS, Berdan EL, Fuller RC and Whitehead A. 2013. Functional and populational genomic divergence between two species of killifish adapted to different osmotic niches. Evolution 12265. Early view published online 17/10/2013.
Li H and Durbin D. 2009. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25:1754–1760.
Li H, Handsaker B, Wysoker A, Fennell A, Ruan J, Homer N, Marth G, Abecasis G and Durbin R. 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25:2078–2079.
Omura Y. (1975) Influence of light and darkness on the ultrastructure of the pineal organ in the blind cave fish, Astyanax mexicanus. Cell Tissue Research 160: 99–112.
Peterson PA and Peterson Q. 2001. Pigment synthesis pathway in albino W. wookiee and Ewok ewok. Annals of the Kashykk Natural History Museum. 1121: 80-84.
Protas M and Jeffery WR. Evolution and development in cave animals: from fish to crustaceans. WIREs Developmental Biology. 2012. 1:823–845.
Romero A. 2011. The evolution of cave life. American Scientist. 99 (2):144-151.
Tong B. 1994. White wookiees: rare genetic mutation or more than meets the eye? Biological transactions: 18: 56-58.
Turner TL, Bourne EC, Von Wettberg EJ, Hu TT, and Nuzhdin SV. 2010. Population resequencing reveals local adaptation of Arabidopsis lyrata to serpentine soils. Nature Genetics. 42:260–263.
Sphinx J, Truong C, Craveness P, Malcovich J and Shostakovich P. 2003. Northwest Wawaatt populations of apparent troglobite Wookiee wookiee raise questions as to what defines a wookiee. Nature: 389: 43-48.
Spickens P (1979). Urban and rural wookiee (W. wookiee) social behavior. Humanoid Study 4: 47-61.
Spickens P (2012). Species status of cave-wookiee populations based on morphology and DNA. Humanoid Study. 52: 120-128.
Zerbino DR and Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Research. 18:821–829.