D.I.Y. P.C.R. NOTES, APPENDUM, SHOOTING THE BREEZE – WHATEVER
(This post will be updated from time to time, as improvements arise with regards to the MAKE magazine thermal cycler project. For more info, go here)
Date: August 25th
We repeated the experiment with the 2C adjustment, but obtained similarily non-optimal data as before. This led to recalibrating of our temperture sensor at 0C and monitoring to see if the temperture cycles waiver at the latter cycles (i.e after the first 10 cycles), which was not the case.
At that point, we decided to calibrate our temperature sensor in real time with the eppendorf cycler to see what actual tempertures were for the reaction. In other words, despite the program being (which represents the block temperature):
STEP 1: 95C 3min
STEP 2: 95C 30sec
STEP 3: 58C 60sec
STEP 4: 72C 90sec
STEP 5: repeat 2 to 4 (x40)
we would actually probe a tube in the Eppendorf machine to see what the real “reaction” temperatures were. In this case, we noted that in reality, our tube was subjected to the following:
STEP 2: 92C 12sec (due to attempted ramp up to the 95C)
STEP 3: 58C 40sec (due to ramp down to 58C)
STEP 4: 72C 70sec (due to ramp up to 72C)
This means, there is the possibility that although the DIY thermal cycler is performing perfectly, the conditions are in fact not congruent with our control samples under the eppendorf machine experiment.
So… current hypotheses to test out:
1. Since we are using el cheapo pTaq, the difference between denaturing at 92C for 12sec versus 95C for 30sec is significant (i.e. higher tempertures is rougher on already poor quality enzyme and is killing it sooner).
2. The difference between the 2C/sec and 1C/sec ramp is significant. We can’t from a kinetic point of view concur with this, but this is testable by redoing experiment using tubes with thinner walls etc.
3. Because the DIY thermal cycler requires oil to prevent evaporation (and our Eppendorf machine doesn’t because it has a heated lid at 105C), somehow the addition of oil is having an effect. Again, unlikely but really easy to test.
4. Trace amounts of the thermal paste in reactions carried out in the DIY thermal cycler is messing things up?
Back to the drawing board…
Date: August 21st, 2006
Current Status:
Presently, we’re still working on optimizing conditions so that visualization via Carolina Blue staining is possible. We’re at a point where data is observable when using standard Ethidium Bromide stains – although the bands are significantly less intense, when compared with data from optimal experimental conditions (i.e. use of a commercially bought PCR machine). We think that due to the almost perfect nature of the temperature profile exhibited by the thermal cycler (see below), there may be an accuracy problem with regards to temperature calibration. i.e. the entire cycle is off by a net amount of degrees centigrade, leading to less efficient amplification (i.e. temperature is most likely a few degrees hotter) (Ed. note: just determined that this is the case, where accuracy was off (temps are hotter) by about 2 to 3oC)
Gel Data
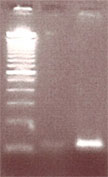
Lane 1: 100bp ladder molecular weight standard, Lane 2: Alu insertion PCR experiment with MAKE DIY thermal cycler, Lane 3: Alu insertion PCR experiment done on an Eppendorf brand thermal cycler. You’ll note one band at 100bp indicative of an -/- genotype.
Anyway, you can see that overall the machine works, but not quite up to par with the Eppendorf control, which we’ll need to meet for a band to be picked up by the less senstive Carolina Blue Stain.
PCR Program
Currently, we’re using the following:
i. 95C 300 sec (this is the hot start – we have an extended one so that we have lots of time to add our reaction at this step – “manual hot start”)
ii. 95C 30sec (denaturation step)
iii. 58C 60sec (annealing of primers step)
iv. 70C 90sec (elongation step – longer to ensure coverage to second primer binding site)
v. repeat step ii to iv (x 40)
Temperature Profile
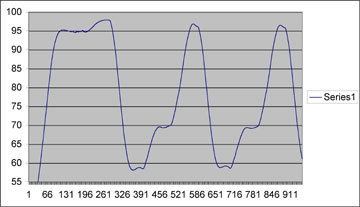
You can see that this is really nice looking, and comparable to commerically bought machine with the main exception of a slower ramping time (time to change from one temperature to the next). Our ramping temperature change rate was at about 0.5C/sec, as compared to the Eppendorf which ramps at about 1C/sec. Note that, as described in the Ed. Notes above, the temperatures on this profile are off by about 3C across the board.
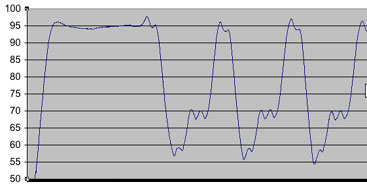
This is a figure of an earlier temperature profile, where you can see a greater wavering of temperatures during the steps within the cycles. This, we were able to correct, by first immersing the heat block/peltier set-up in a tupperware container filled with water that was prewarmed to approximately 60C. As well, although initially, we monitored temperature with a tube filled with about 50ul volume worth of thermal paste, we noted that it’s important for the temperature monitor to be reading from a scenario similar to the reaction (i.e. about 50ul of fluid, plus a few drops of mineral oil).
Heat Stable Polymerase
We’re still using a pTaq polymerase, which actually would be significantly less efficient than the Invitrogen enzyme suggested in the text. However, because we’re still in troubleshooting mode, we thought it best to play with an enzyme that we have a large supply of. Regardless, pTaq is working well with our control (Eppendorf brand machine).