GENETIC STUDIES OF AGING AND LONGEVITY IN MODEL ORGANISMS
(August 2004)
Aging can be characterized as:
(1) an inevitable consequence of being a multicellular organism;
(2) associated with a random, passive decline in function;
(3) leading to a global loss of homeostasis (the state of sustained equilibrium in which all cells, and all life forms, exist) over time; and
(4) mortality increasing with age [1].
Age is not a disease, but it does predispose an organism to a variety of diseases [2], in the case of humans, this includes heart disease, arthritis, osteoporosis, diabetes, cancer and Alzheimer’s.
Achieving longevity is thought to be caused by a combination of factors. Possessing longevity-enabling genes, lacking disease-predisposing genetic variations and appropriate health-related behavior are all thought to contribute to long life [3] . Therefore, understanding of the genetics of aging and longevity should lead to the discovery of genes and ultimately drugs that slow down the aging process and facilitate people’s ability to delay and perhaps escape age-associated diseases.
Humans’ relatively long life expectancy makes them less attractive as subjects for longevity research. Using short-lived organisms to discover life-span-altering genes is much more feasible. This paper describes the current state of aging and longevity research with model organisms.
Genetic Model Systems
The major genetic model systems used in aging research are those of the filamentous fungus Podospora anserine, baker’s yeast Saccharomyces cerevisiae, the roundworm Canenorhabditis elegans (C. elegans), the fruit fly Drosophila melanogaster (D. melanogaster), and the common mouse.
These model systems have many advantages for genetic dissection of aging and longevity mechanisms. They are small, easy to maintain, and have a short life span. For instance, life cycle of yeasts and worms is only few days. This allows experiments to be concluded within weeks of their initiation. In addition, there is significant homology between the human genome and those of simpler organisms. About one-fifth of human disease genes that have been positionally cloned possess yeast homologues, reflecting a remarkable degree of genetic conservation [4].
Several genetic methodologies have been used to study aging in these organisms [2]. Mutagenesis is a method that affects the expression of genes or the structure of gene products by altering of DNA sequences using physical or chemical treatment, followed by screening for individuals that possess the resultant phenotypes (visible characteristics and/or behavior that result from the interaction of an organism’s genotype and the environment). Molecular genetics strategies are similar except that they target known genetic loci that are thought to be involved in expressing the phenotype of interest. Quantitative trait loci (QTL) analysis encompasses a family of techniques that are used to zero in on the location of genes responsible for the variation in a particular phenotype. Selective breeding is used to progressively comb together the genetic variants in a population that are responsible for a particular trait by choosing individuals with the most extreme phenotypic expression for breeding. These genetic methodologies have now identified 35 cloned genes that determine longevity in model organisms. These genes encode a wide variety of proteins with a variety of functions, but they can be categorized into four broad physiological processes that play a role in aging: metabolic control, resistance to stress, gene dysregulation, and genetic stability. Insulin is the major hormone controlling critical energy functions. Below is an example of an insulin-like signaling pathway that plays a role in the control of metabolism, development and longevity in C. elegans.
Insulin-like signaling pathway
Studies of long-lived mutants of the nematode C. elegans have contributed significantly to the progress made in the study of aging during the last decade. An example of what has been learned is the discovery of an insulin-like signaling pathway that regulates longevity and metabolism in C. elegans [5].
![insulinlikepath[2]-1.gif](https://www.scq.ubc.ca/wp-content/uploads/2006/07/insulinlikepath[2]-1.gif)
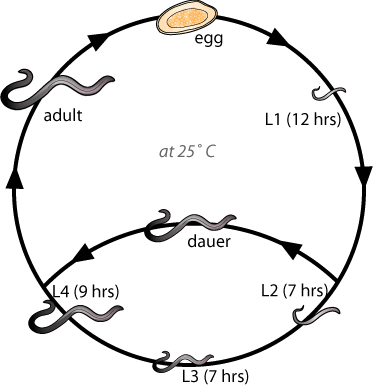
After embryonic development and hatching, the worms move through a series of four larval stages (L1-L4) before reaching adulthood. If they encounter particular environmental stress, worms will enter the dauer larval stage, which allows worms to survive adverse conditions. When favourable environmental conditions arise, dauer larvae resume development and proceed to L4 larval stage and subsequently to adulthood (Figure 2). In dauer state, worms can survive up to 6 months, which is much longer than adult worm life span (2-3 weeks). The increase in lifespan of the dauer state is not entirely due to reduced metabolism, but also due to increased levels of antioxidant enzyme and stress-resistance proteins [6]. The insulin-like pathway is part of a global endocrine system that controls whether the worms grow reproductively or arrest at the dauer stage.
During the time of dauer formation, numerous dauer formation (daf) genes are active and some of them play roles in determining the two alternative developmental fates (dauer or L3 stage) of C. elegans. These regulatory genes were identified by two general classes of mutants: dauer defective and dauer constitutive mutants. Mutants that possess Dauer constitutive genes such as daf-2 and age-1 which constitute components of the C. elegans insulin signaling pathway live 2 to 3 times longer than wild type [8-10]. Conversely, daf-16 dauer defective mutants suppress the increase in longevity caused by daf-2 and age-1 mutants. Based on previous genetic studies, several dauer genes have been ordered into an insulin-like signaling pathway [5,11-15]. Under reproductive growth conditions, high daf-2 receptor signaling activates age-1 and pdk-1 which in turn, activates the akt-1 and -2 kinases, which negatively regulate daf-16 activity. Inactivated daf-16 fails to further activate the genes necessary for dauer arrest and long life span or repress the genes that inhibit reproductive growth and short life span [7]. Under dauer inducing conditions, these kinase cascades are inactive, so then active daf-16 represses genes required for reproductive growth and short life span, and/or activate genes necessary for dauer arrest and long life span [7]. (Figure 1)
In addition to metabolism, the C. elegans insulin pathway regulates free radical protection [16]. Moreover, the insulin pathway in worm aging may be related to the longevity increase caused by caloric restriction in mammals [17]. Caloric restriction in mammals may cause a decline in insulin-like signaling that induces a partial diapause state [18], like that induced in weak daf-2 and age-1 mutants.
Recently, two research group described mutants of genes that modulate pathways homologous to insulin signaling pathways in humans and increase life span of D. melanogaster. A mutation of the insulin-like receptor gene [19], which is homologous to the C. elegans daf-2 gene, and a mutation of the D. melanogaster gene chico 20, which participates in an insulin-like signaling pathway, are associated with a significant increase in life span. Thus, the role of insulin-like signaling pathways appears to be evolutionarily conserved in a number of species, suggesting the importance of investigating such pathways and their relation to aging and longevity in humans.
Conclusion
Genetic studies in simpler organisms demonstrate that specific genes have potent influences on life span. These findings provide a framework for understanding the factors by which life span is determined in lower organisms. More importantly, organism models provide interesting phenotypic characteristics and candidate pathways to be studied in relation to longevity, such as parameters of metabolic control, stress resistance and genetic instability. However, investigation of human homologues of these genes would naturally be more complicated because aging and susceptibility to diseases associated with aging in human is influenced by multiple genes as well as environment factors.
Glossary
1. Mutagenesis – The production of either random or specific mutations in a piece of cloned DNA. Typically, the DNA will then be reintroduced into a cell or an organism to assess the results of the mutagenesis.
2. Quantitative Trait Loci – Continuous traits are often measured and given a quantitative value. Therefore, they are often referred to as quantitative traits, and the area of genetics that studies their mode of inheritance is called quantitative genetics. Many human phenotypes such as IQ, learning ability and blood pressure are quantitative traits. These traits are controlled by multiple genes, each segregating according to Mendel’s laws.
3. Metabolic Control – Mechanisms by which the chemical reactions that occur in a living cell are controlled.
References
1. Helfand, S.L. & Rogina, B. Molecular genetics of aging in the fly: is this the end of the beginning? Bioessays 25, 134-41 (2003).
2. Jazwinski, S.M. Aging and longevity genes. Acta Biochim Pol 47, 269-79 (2000).
3.Perls, T., Kunkel, L. & Puca, A. The genetics of aging. Curr Opin Genet Dev 12, 362-9 (2002).
4. Bassett, D.E., Jr. et al. Genome cross-referencing and XREFdb: implications for the identification and analysis of genes mutated in human disease. Nat Genet 15, 339-44 (1997).
5. Kimura, K.D., Tissenbaum, H.A., Liu, Y. & Ruvkun, G. daf-2, an insulin receptor-like gene that regulates longevity and diapause in Caenorhabditis elegans. Science 277, 942-6 (1997).
6.Murakami, S. & Johnson, T.E. A genetic pathway conferring life extension and resistance to UV stress in Caenorhabditis elegans. Genetics 143, 1207-18 (1996).
7. Finch, C.E. & Ruvkun, G. The genetics of aging. Annu Rev Genomics Hum Genet 2, 435-62 (2001).
8. Dorman, J.B., Albinder, B., Shroyer, T. & Kenyon, C. The age-1 and daf-2 genes function in a common pathway to control the lifespan of Caenorhabditis elegans. Genetics 141, 1399-406 (1995).
9. Kenyon, C., Chang, J., Gensch, E., Rudner, A. & Tabtiang, R. A C. elegans mutant that lives twice as long as wild type. Nature 366, 461-4 (1993).
10. Larsen, P.L., Albert, P.S. & Riddle, D.L. Genes that regulate both development and longevity in Caenorhabditis elegans. Genetics 139, 1567-83 (1995).
11. Morris, J.Z., Tissenbaum, H.A. & Ruvkun, G. A phosphatidylinositol-3-OH kinase family member regulating longevity and diapause in Caenorhabditis elegans. Nature 382, 536-9 (1996).
12. Ogg, S. et al. The Fork head transcription factor DAF-16 transduces insulin-like metabolic and longevity signals in C. elegans. Nature 389, 994-9 (1997).
13. Ogg, S. & Ruvkun, G. The C. elegans PTEN homolog, DAF-18, acts in the insulin receptor-like metabolic signaling pathway. Mol Cell 2, 887-93 (1998).
14. Paradis, S. & Ruvkun, G. Caenorhabditis elegans Akt/PKB transduces insulin receptor-like signals from AGE-1 PI3 kinase to the DAF-16 transcription factor. Genes Dev 12, 2488-98 (1998).
15. Paradis, S., Ailion, M., Toker, A., Thomas, J.H. & Ruvkun, G. A PDK1 homolog is necessary and sufficient to transduce AGE-1 PI3 kinase signals that regulate diapause in Caenorhabditis elegans. Genes Dev 13, 1438-52 (1999).
16. Taub, J. et al. A cytosolic catalase is needed to extend adult lifespan in C. elegans daf-C and clk-1 mutants. Nature 399, 162-6 (1999).
17. Sohal, R.S. & Weindruch, R. Oxidative stress, caloric restriction, and aging. Science 273, 59-63 (1996).
18. Tauber MJ, T.C., Masaki S. Seasonal Adaptation of Insects., 411pp. (Oxford Univ. Press, New York, 1986).
19. Tatar, M. et al. A mutant Drosophila insulin receptor homolog that extends life-span and impairs neuroendocrine function. Science 292, 107-10 (2001).
20. Clancy, D.J. et al. Extension of life-span by loss of CHICO, a Drosophila insulin receptor substrate protein. Science 292, 104-6 (2001).
(Art by Jen Philpot)