A HISTORY It began, as all things do, with a geology joke. We ranked candy based on their ability to elicit “joy” or “despair” and then worked in that ranking in various geological strata, both real and imagined. The strata, not the ranking. This started in 2006, and grew from there. At the beginning, we…
The Science Creative Quarterly
From journal club
Reductive Genome Evolution of Obligate Symbiont Midichloria humanum: Implications of Gene Loss
(Click on the front page image to download the full article) Abstract: We sequenced and analyzed the genome of the Midichloria humanum, a ubiquitous intracellular symbiont, in 34 human subjects covering a range of population sizes from 1,000 to over 14,000 midichlorians per human cell. Midichlorians mediate the relationship between their human hosts and the Force, which confers exceptional physical and cognitive abilities on hosts with per-cell midichlorian populations (‘midichlorian count,’ mdc) larger than 10,000. Although, quantitatively, the genome of M. humanum is more extensively degraded than those of closely related Rickettsia species, midichlorians retained many intact genes involved in…
Expression Changes of Genes Associated With Structural Proteins May Be Related to Adaptive Skin Characteristics Specific to Metamorphmagi
(Click on the front page image to download the full article) Abstract: Metamorphmagi have the ability to change their physical characteristics at will, without requiring the use of potions or spells. As such, metamorphmagi skin contains morphologically and functionally distinct properties from non-metamorphmagi skin, providing metamorphagi with heightened skin flexibility and durability. However, previous studies that have examined metamorphmagi are largely qualitative or anecdotal. Thus, we aimed to quantitatively compare metamorphmagi skin to non-metamorphmagi skin and examine the genetic causes underlying metamorphmagi-specific skin characteristics.. We performed histology on cross-sections of skin and quantitatively demonstrated that the epidermis and dermis of…
The Unicorn Microbiome: A proposed microbial mechanism for the rainbow pigments in unicorn poop.
(Click on the front page image to download the full article) Summary: Unicorns (Equus unicornis) are a rare terrestrial mammal, which have been noted to produce a variegated pigmented feces that is often referred to as “rainbow poop.” Despite considerable commercial interest in unicorn rainbow poop, the mechanism leading to this rainbow-pigmentation remains unknown. Microbial metabolism—including the production of pigments— can influence the perceived color of animal excreta. To determine if the rainbow pigments of unicorn feces could have a microbial origin, we used a bioinformatic and microbial cultivation-based approach. To establish what microorganisms could be producing such colored pigments,…
Genetic Association between WZRD-1 Variants and the Variation in Magical Abilities in Canadians
(Click on the front page image to download the full article) Abstract: Current literature lacks a consensus on the mechanism of biological inheritance of magical abilities. Past findings have suggested that the presence of restriction enzyme digestion sites in WZRD-1 are associated with variations in human magical abilities. In this study, Next Generation Sequencing (NGS) was performed to characterize the genetic sequence of WZRD-1 in 610 Canadian subjects. These individuals were divided into three cohorts as determined by their magical abilities and blood status. Four genetic variants within WZRD-1 were identified, and the genotypic and allelic frequencies were determined for…
MUTATIONS IN THE SEED REGION OF WIZARD mIR-96 ARE RESPONSIBLE FOR HEREDITARY PARSELTONGUE HYPERACUSIS
Annals of Praetachoral Mechanics (2016). Vol 2. Advanced online publication. download pdf ABSTRACT Parseltongue is a rare hereditary trait that allows wizards to communicate in the language of serpents. Wizards who are parseltongue not only have the ability to speak to serpents (parselmouth), but also possess the ability to hear ultrasonic frequencies produced by the snake (parseltongue hyperacusis). Genome wide association studies (GWAS) have identified scores of genetic variants that contribute to different wizarding phenomenon. MicroRNAs negatively regulate mRNA expression by binding complementary sites in target mRNAs, with the specificity of interaction being crucially dependent on the miRNA seed region.…
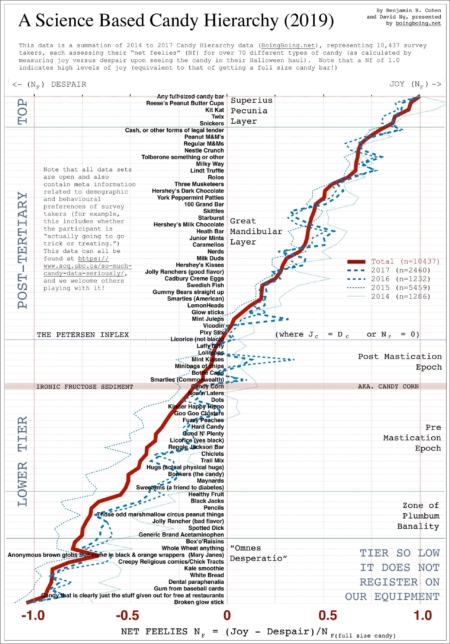
WHY SOME PEOPLE MAKE THE WRONG DECISION AND OTHERS DON’T: AN ASSESSMENT OF ARITHMETIC AND GEOMETRIC PROJECTIONS OF THE FRIDAY-SUNDAY DIVIDE BEYOND THE MEHSPAIR CHASM WITH P-VALUES, χ2, AND LICORICE
ABSTRACT So it’s come to this. Another year has passed and another Candy Hierarchy survey has been completed. Check that. “Candy Hierarchy” is what yahoos call it, but to us, it’s a deep dive into the Friday/Sunday question that has blessed our species since caveman times. Caveman times. We’ve taken a look at the data, and frankly, we don’t like what we see. Maybe it’s not all bad. Maybe. Let’s start with the good. Remember last year when a couple of plucky scientists who had overcome obscure forms of childhood adversity before finding their footing in a ne’er do well…
2016 CANDY HIERARCHY: SUPPLEMENTAL RAW DATA AND NEW ANALYSES
PREAMBLE: This is for you data nerds who might want to dig into the data a little deeper (we didn’t have a lot of time to play around with it). We’ve provided access to the full excel spreadsheet (which also includes data that was cut off from our analysis due to us having to get started earlier for today’s piece on BoingBoing). We’re essentially hoping that with more rigorous analysis of the data, we might be able to find out whether there are statistically relevant differences in how different folks rate their candies For example, do people who see the…
NucSh REGULATES NECESSARY NUCLEIC SHRINKAGE FOR GENOME SWAP IN POLYJUICE POTION
Annals of Praetachoral Mechanics (2016). Vol 2. Advanced online publication. download pdf ABSTRACT This study provides further investigations into the wizarding potion known commonly as the Polyjuice potion; and specifically to assess the contribution of the known fluxweed protein, NucSh. NucSh has been previously shown, via co-immunoprecipitation experiments, to be part of the PhageCrux Complex, which in turn appears to be key for many major transformative magic related properties. Other studies have also shown that the principle factor of this Auror favorite is possibly related to a symbiotically absorbed and retained bacteriophage present in Lacewing flies (Borage et al, 2009).…